Sarfaraz Hussein
Joint Stream: Malignant Region Learning for Breast Cancer Diagnosis
Jun 26, 2024



Abstract:Early diagnosis of breast cancer (BC) significantly contributes to reducing the mortality rate worldwide. The detection of different factors and biomarkers such as Estrogen receptor (ER), Progesterone receptor (PR), Human epidermal growth factor receptor 2 (HER2) gene, Histological grade (HG), Auxiliary lymph node (ALN) status, and Molecular subtype (MS) can play a significant role in improved BC diagnosis. However, the existing methods predict only a single factor which makes them less suitable to use in diagnosis and designing a strategy for treatment. In this paper, we propose to classify the six essential indicating factors (ER, PR, HER2, ALN, HG, MS) for early BC diagnosis using H\&E stained WSI's. To precisely capture local neighboring relationships, we use spatial and frequency domain information from the large patch size of WSI's malignant regions. Furthermore, to cater the variable number of regions of interest sizes and give due attention to each region, we propose a malignant region learning attention network. Our experimental results demonstrate that combining spatial and frequency information using the malignant region learning module significantly improves multi-factor and single-factor classification performance on publicly available datasets.
Automated Segmentation and Recurrence Risk Prediction of Surgically Resected Lung Tumors with Adaptive Convolutional Neural Networks
Sep 17, 2022
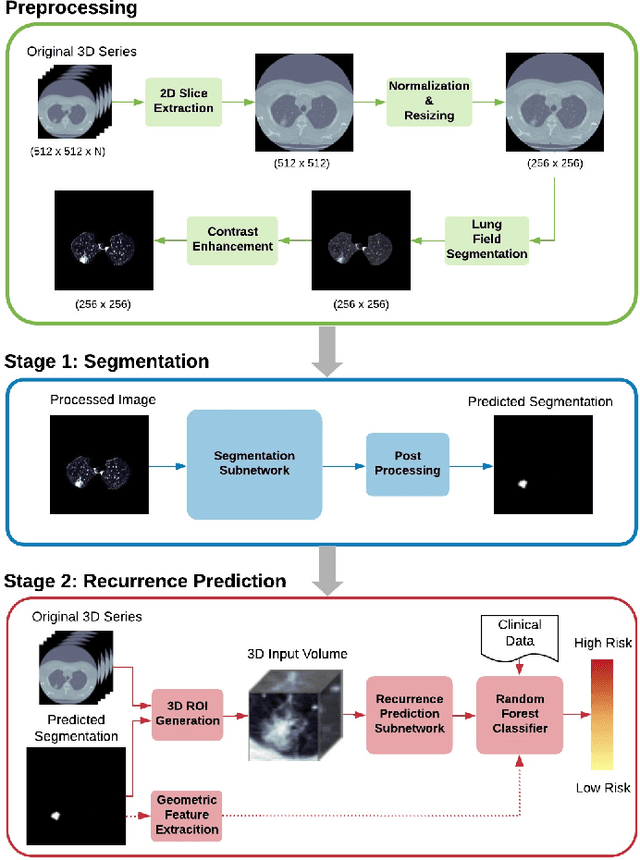
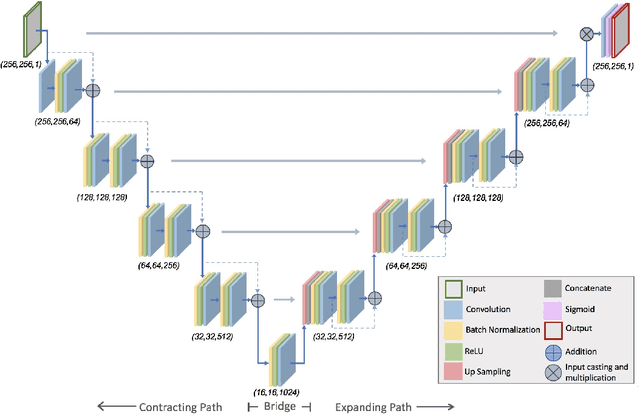
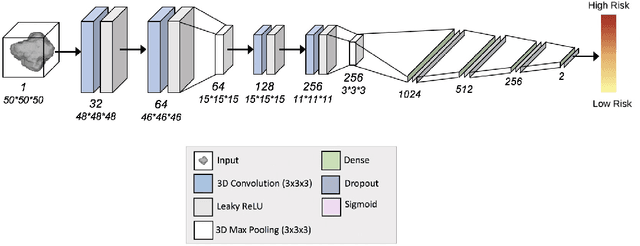
Abstract:Lung cancer is the leading cause of cancer related mortality by a significant margin. While new technologies, such as image segmentation, have been paramount to improved detection and earlier diagnoses, there are still significant challenges in treating the disease. In particular, despite an increased number of curative resections, many postoperative patients still develop recurrent lesions. Consequently, there is a significant need for prognostic tools that can more accurately predict a patient's risk for recurrence. In this paper, we explore the use of convolutional neural networks (CNNs) for the segmentation and recurrence risk prediction of lung tumors that are present in preoperative computed tomography (CT) images. First, expanding upon recent progress in medical image segmentation, a residual U-Net is used to localize and characterize each nodule. Then, the identified tumors are passed to a second CNN for recurrence risk prediction. The system's final results are produced with a random forest classifier that synthesizes the predictions of the second network with clinical attributes. The segmentation stage uses the LIDC-IDRI dataset and achieves a dice score of 70.3%. The recurrence risk stage uses the NLST dataset from the National Cancer institute and achieves an AUC of 73.0%. Our proposed framework demonstrates that first, automated nodule segmentation methods can generalize to enable pipelines for a wide range of multitask systems and second, that deep learning and image processing have the potential to improve current prognostic tools. To the best of our knowledge, it is the first fully automated segmentation and recurrence risk prediction system.
Out of distribution detection for skin and malaria images
Nov 02, 2021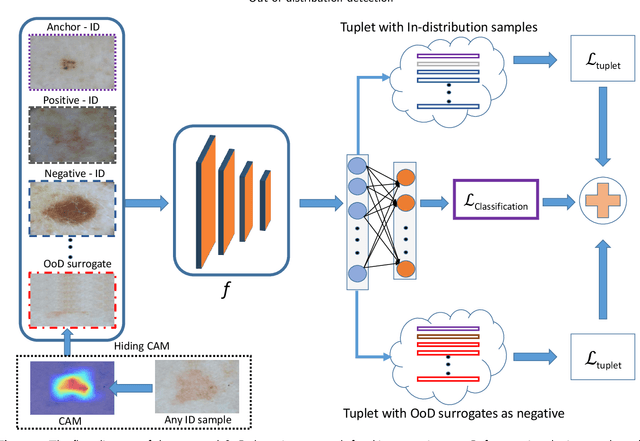
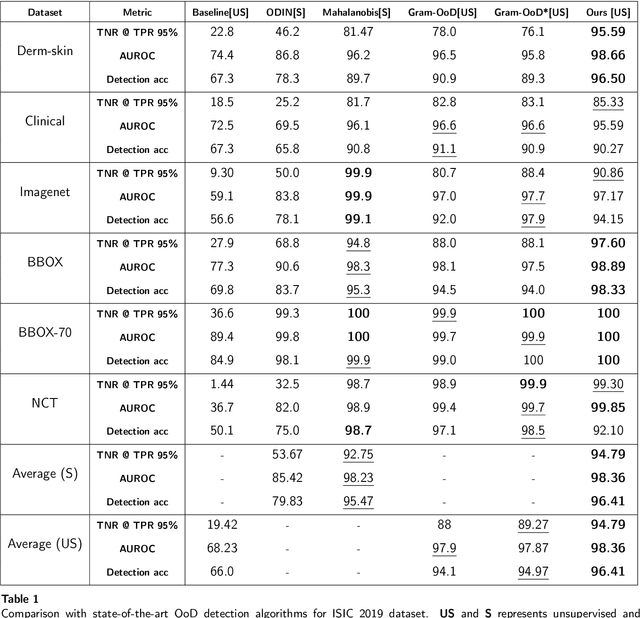
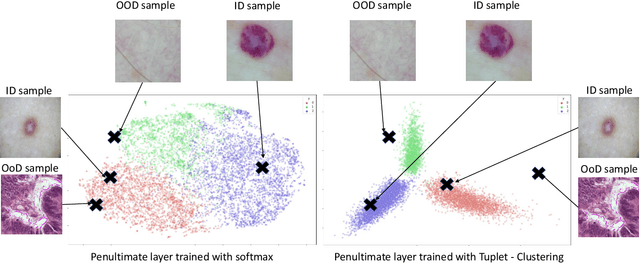
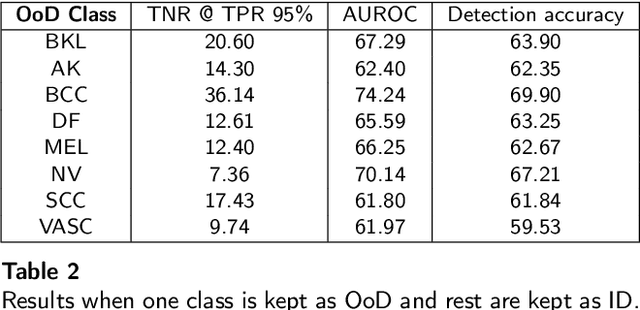
Abstract:Deep neural networks have shown promising results in disease detection and classification using medical image data. However, they still suffer from the challenges of handling real-world scenarios especially reliably detecting out-of-distribution (OoD) samples. We propose an approach to robustly classify OoD samples in skin and malaria images without the need to access labeled OoD samples during training. Specifically, we use metric learning along with logistic regression to force the deep networks to learn much rich class representative features. To guide the learning process against the OoD examples, we generate ID similar-looking examples by either removing class-specific salient regions in the image or permuting image parts and distancing them away from in-distribution samples. During inference time, the K-reciprocal nearest neighbor is employed to detect out-of-distribution samples. For skin cancer OoD detection, we employ two standard benchmark skin cancer ISIC datasets as ID, and six different datasets with varying difficulty levels were taken as out of distribution. For malaria OoD detection, we use the BBBC041 malaria dataset as ID and five different challenging datasets as out of distribution. We achieved state-of-the-art results, improving 5% and 4% in TNR@TPR95% over the previous state-of-the-art for skin cancer and malaria OoD detection respectively.
Estimation of BMI from Facial Images using Semantic Segmentation based Region-Aware Pooling
Apr 10, 2021

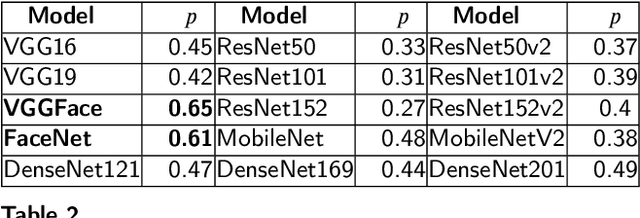

Abstract:Body-Mass-Index (BMI) conveys important information about one's life such as health and socio-economic conditions. Large-scale automatic estimation of BMIs can help predict several societal behaviors such as health, job opportunities, friendships, and popularity. The recent works have either employed hand-crafted geometrical face features or face-level deep convolutional neural network features for face to BMI prediction. The hand-crafted geometrical face feature lack generalizability and face-level deep features don't have detailed local information. Although useful, these methods missed the detailed local information which is essential for exact BMI prediction. In this paper, we propose to use deep features that are pooled from different face regions (eye, nose, eyebrow, lips, etc.,) and demonstrate that this explicit pooling from face regions can significantly boost the performance of BMI prediction. To address the problem of accurate and pixel-level face regions localization, we propose to use face semantic segmentation in our framework. Extensive experiments are performed using different Convolutional Neural Network (CNN) backbones including FaceNet and VGG-face on three publicly available datasets: VisualBMI, Bollywood and VIP attributes. Experimental results demonstrate that, as compared to the recent works, the proposed Reg-GAP gives a percentage improvement of 22.4\% on VIP-attribute, 3.3\% on VisualBMI, and 63.09\% on the Bollywood dataset.
Deep Detector Health Management under Adversarial Campaigns
Nov 19, 2019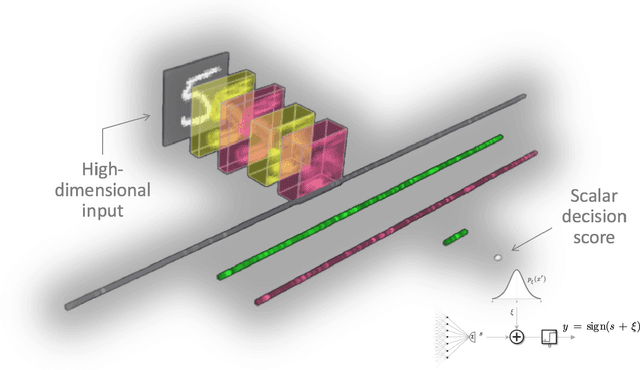
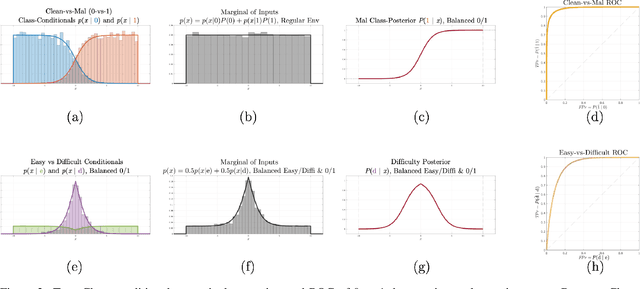
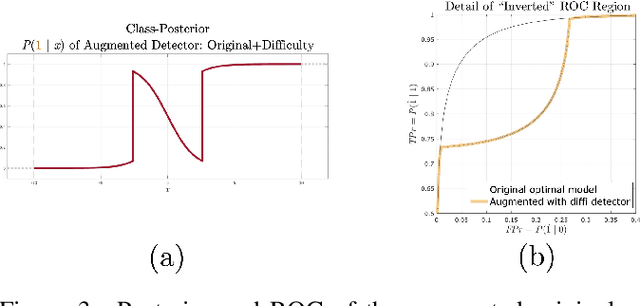
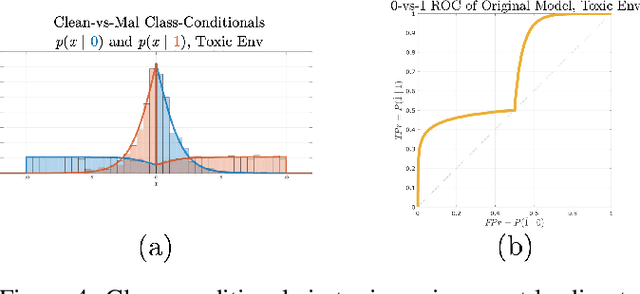
Abstract:Machine learning models are vulnerable to adversarial inputs that induce seemingly unjustifiable errors. As automated classifiers are increasingly used in industrial control systems and machinery, these adversarial errors could grow to be a serious problem. Despite numerous studies over the past few years, the field of adversarial ML is still considered alchemy, with no practical unbroken defenses demonstrated to date, leaving PHM practitioners with few meaningful ways of addressing the problem. We introduce turbidity detection as a practical superset of the adversarial input detection problem, coping with adversarial campaigns rather than statistically invisible one-offs. This perspective is coupled with ROC-theoretic design guidance that prescribes an inexpensive domain adaptation layer at the output of a deep learning model during an attack campaign. The result aims to approximate the Bayes optimal mitigation that ameliorates the detection model's degraded health. A proactively reactive type of prognostics is achieved via Monte Carlo simulation of various adversarial campaign scenarios, by sampling from the model's own turbidity distribution to quickly deploy the correct mitigation during a real-world campaign.
A Novel Extension to Fuzzy Connectivity for Body Composition Analysis: Applications in Thigh, Brain, and Whole Body Tissue Segmentation
Oct 14, 2018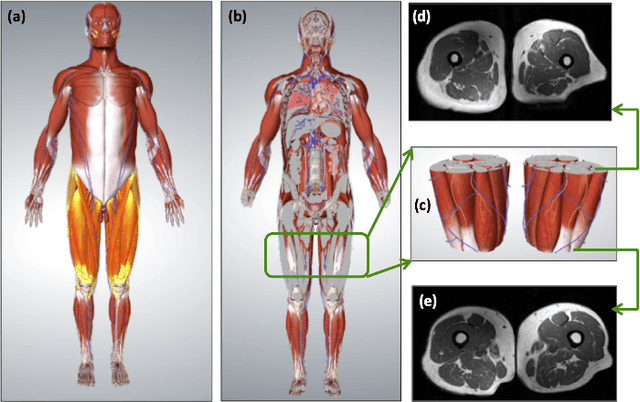
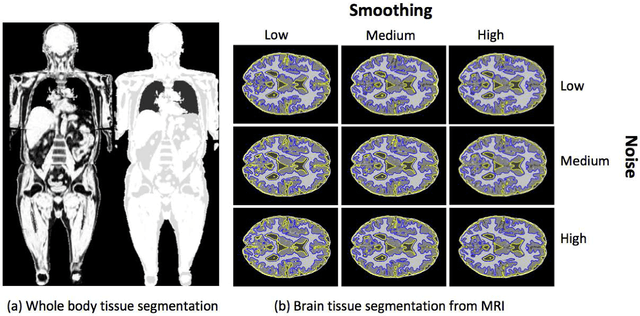
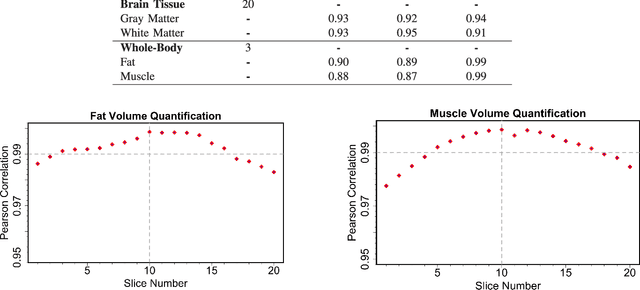
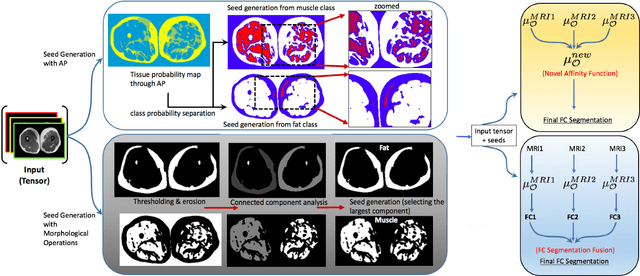
Abstract:Magnetic resonance imaging (MRI) is the non-invasive modality of choice for body tissue composition analysis due to its excellent soft tissue contrast and lack of ionizing radiation. However, quantification of body composition requires an accurate segmentation of fat, muscle and other tissues from MR images, which remains a challenging goal due to the intensity overlap between them. In this study, we propose a fully automated, data-driven image segmentation platform that addresses multiple difficulties in segmenting MR images such as varying inhomogeneity, non-standardness, and noise, while producing high-quality definition of different tissues. In contrast to most approaches in the literature, we perform segmentation operation by combining three different MRI contrasts and a novel segmentation tool which takes into account variability in the data. The proposed system, based on a novel affinity definition within the fuzzy connectivity (FC) image segmentation family, prevents the need for user intervention and reparametrization of the segmentation algorithms. In order to make the whole system fully automated, we adapt an affinity propagation clustering algorithm to roughly identify tissue regions and image background. We perform a thorough evaluation of the proposed algorithm's individual steps as well as comparison with several approaches from the literature for the main application of muscle/fat separation. Furthermore, whole-body tissue composition and brain tissue delineation were conducted to show the generalization ability of the proposed system. This new automated platform outperforms other state-of-the-art segmentation approaches both in accuracy and efficiency.
Supervised and Unsupervised Tumor Characterization in the Deep Learning Era
Jul 29, 2018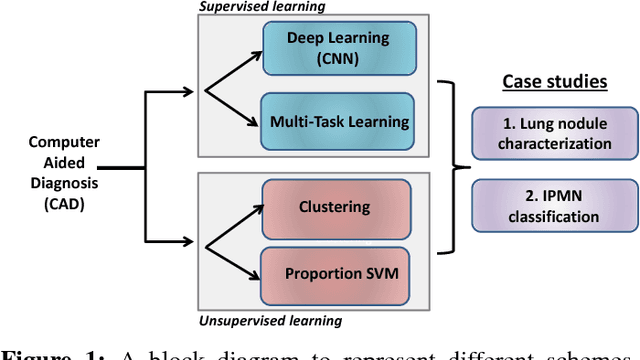

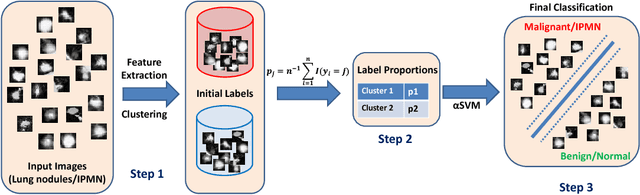
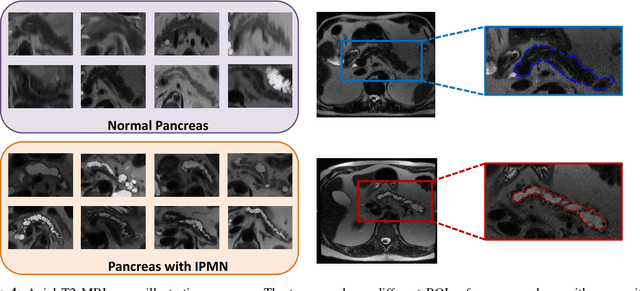
Abstract:Cancer is among the leading causes of death worldwide. Risk stratification of cancer tumors in radiology images can be improved with computer-aided diagnosis (CAD) tools which can be made faster and more accurate. Tumor characterization through CADs can enable non-invasive cancer staging and prognosis, and foster personalized treatment planning as a part of precision medicine. In this study, we propose both supervised and unsupervised machine learning strategies to improve tumor characterization. Our first approach is based on supervised learning for which we demonstrate significant gains in deep learning algorithms, particularly by utilizing a 3D Convolutional Neural Network along with transfer learning. Motivated by the radiologists' interpretations of the scans, we then show how to incorporate task dependent feature representations into a CAD system via a "graph-regularized sparse Multi-Task Learning (MTL)" framework. In the second approach, we explore an unsupervised scheme to address the limited availability of labeled training data, a common problem in medical imaging applications. Inspired by learning from label proportion (LLP) approaches, we propose a new algorithm, proportion-SVM, to characterize tumor types. We also seek the answer to the fundamental question about the goodness of "deep features" for unsupervised tumor classification. We evaluate our proposed approaches (both supervised and unsupervised) on two different tumor diagnosis challenges: lung and pancreas with 1018 CT and 171 MRI scans respectively.
Deep Multi-Modal Classification of Intraductal Papillary Mucinous Neoplasms (IPMN) with Canonical Correlation Analysis
Apr 27, 2018
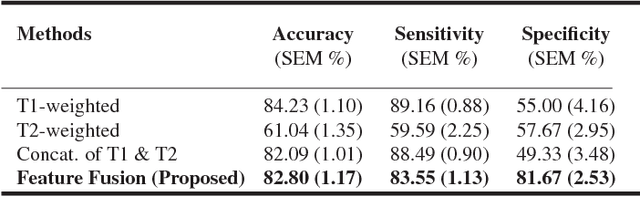
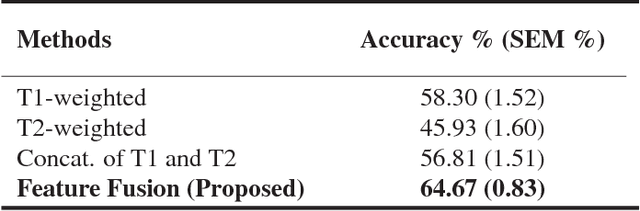
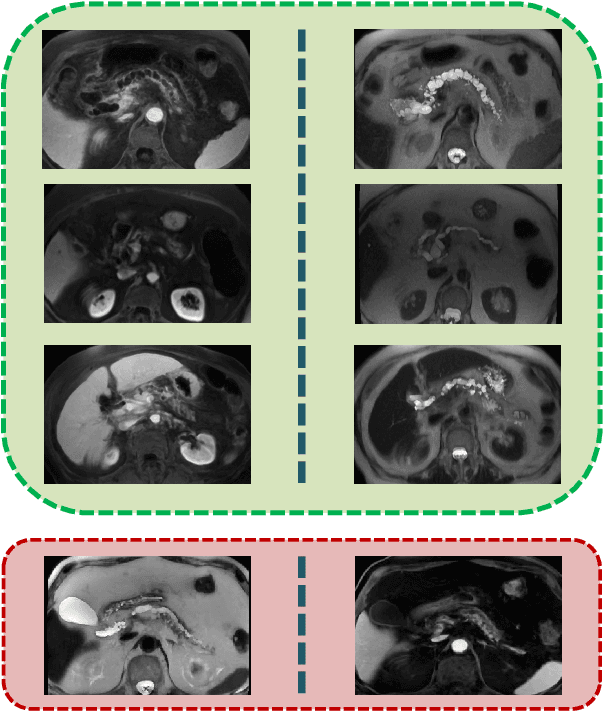
Abstract:Pancreatic cancer has the poorest prognosis among all cancer types. Intraductal Papillary Mucinous Neoplasms (IPMNs) are radiographically identifiable precursors to pancreatic cancer; hence, early detection and precise risk assessment of IPMN are vital. In this work, we propose a Convolutional Neural Network (CNN) based computer aided diagnosis (CAD) system to perform IPMN diagnosis and risk assessment by utilizing multi-modal MRI. In our proposed approach, we use minimum and maximum intensity projections to ease the annotation variations among different slices and type of MRIs. Then, we present a CNN to obtain deep feature representation corresponding to each MRI modality (T1-weighted and T2-weighted). At the final step, we employ canonical correlation analysis (CCA) to perform a fusion operation at the feature level, leading to discriminative canonical correlation features. Extracted features are used for classification. Our results indicate significant improvements over other potential approaches to solve this important problem. The proposed approach doesn't require explicit sample balancing in cases of imbalance between positive and negative examples. To the best of our knowledge, our study is the first to automatically diagnose IPMN using multi-modal MRI.
How to Fool Radiologists with Generative Adversarial Networks? A Visual Turing Test for Lung Cancer Diagnosis
Jan 09, 2018



Abstract:Discriminating lung nodules as malignant or benign is still an underlying challenge. To address this challenge, radiologists need computer aided diagnosis (CAD) systems which can assist in learning discriminative imaging features corresponding to malignant and benign nodules. However, learning highly discriminative imaging features is an open problem. In this paper, our aim is to learn the most discriminative features pertaining to lung nodules by using an adversarial learning methodology. Specifically, we propose to use unsupervised learning with Deep Convolutional-Generative Adversarial Networks (DC-GANs) to generate lung nodule samples realistically. We hypothesize that imaging features of lung nodules will be discriminative if it is hard to differentiate them (fake) from real (true) nodules. To test this hypothesis, we present Visual Turing tests to two radiologists in order to evaluate the quality of the generated (fake) nodules. Extensive comparisons are performed in discerning real, generated, benign, and malignant nodules. This experimental set up allows us to validate the overall quality of the generated nodules, which can then be used to (1) improve diagnostic decisions by mining highly discriminative imaging features, (2) train radiologists for educational purposes, and (3) generate realistic samples to train deep networks with big data.
Risk Stratification of Lung Nodules Using 3D CNN-Based Multi-task Learning
Apr 28, 2017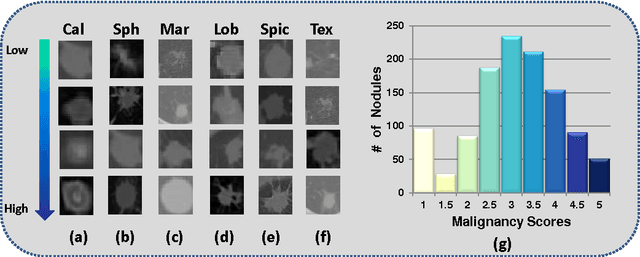

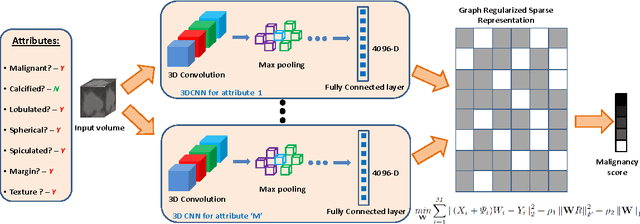
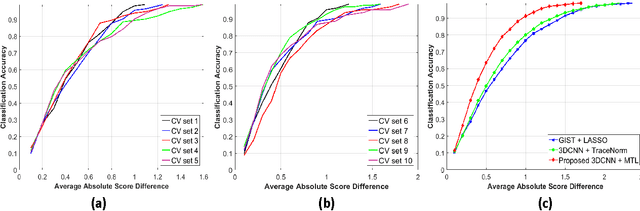
Abstract:Risk stratification of lung nodules is a task of primary importance in lung cancer diagnosis. Any improvement in robust and accurate nodule characterization can assist in identifying cancer stage, prognosis, and improving treatment planning. In this study, we propose a 3D Convolutional Neural Network (CNN) based nodule characterization strategy. With a completely 3D approach, we utilize the volumetric information from a CT scan which would be otherwise lost in the conventional 2D CNN based approaches. In order to address the need for a large amount for training data for CNN, we resort to transfer learning to obtain highly discriminative features. Moreover, we also acquire the task dependent feature representation for six high-level nodule attributes and fuse this complementary information via a Multi-task learning (MTL) framework. Finally, we propose to incorporate potential disagreement among radiologists while scoring different nodule attributes in a graph regularized sparse multi-task learning. We evaluated our proposed approach on one of the largest publicly available lung nodule datasets comprising 1018 scans and obtained state-of-the-art results in regressing the malignancy scores.
* Accepted for publication at Information Processing in Medical Imaging (IPMI) 2017
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge