Jeremy Burt
How to Fool Radiologists with Generative Adversarial Networks? A Visual Turing Test for Lung Cancer Diagnosis
Jan 09, 2018



Abstract:Discriminating lung nodules as malignant or benign is still an underlying challenge. To address this challenge, radiologists need computer aided diagnosis (CAD) systems which can assist in learning discriminative imaging features corresponding to malignant and benign nodules. However, learning highly discriminative imaging features is an open problem. In this paper, our aim is to learn the most discriminative features pertaining to lung nodules by using an adversarial learning methodology. Specifically, we propose to use unsupervised learning with Deep Convolutional-Generative Adversarial Networks (DC-GANs) to generate lung nodule samples realistically. We hypothesize that imaging features of lung nodules will be discriminative if it is hard to differentiate them (fake) from real (true) nodules. To test this hypothesis, we present Visual Turing tests to two radiologists in order to evaluate the quality of the generated (fake) nodules. Extensive comparisons are performed in discerning real, generated, benign, and malignant nodules. This experimental set up allows us to validate the overall quality of the generated nodules, which can then be used to (1) improve diagnostic decisions by mining highly discriminative imaging features, (2) train radiologists for educational purposes, and (3) generate realistic samples to train deep networks with big data.
Multi-Planar Deep Segmentation Networks for Cardiac Substructures from MRI and CT
Aug 03, 2017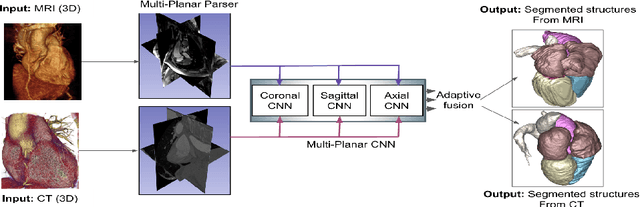
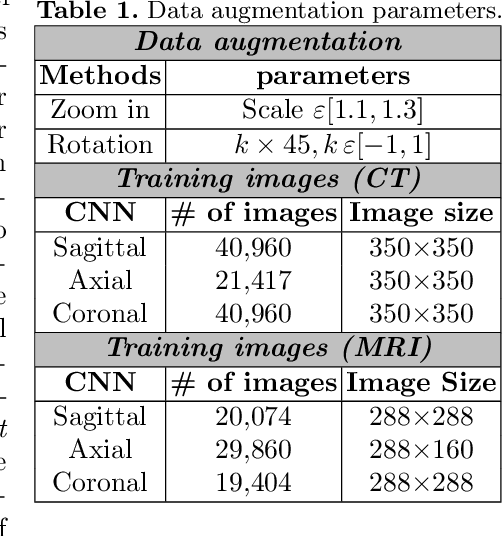
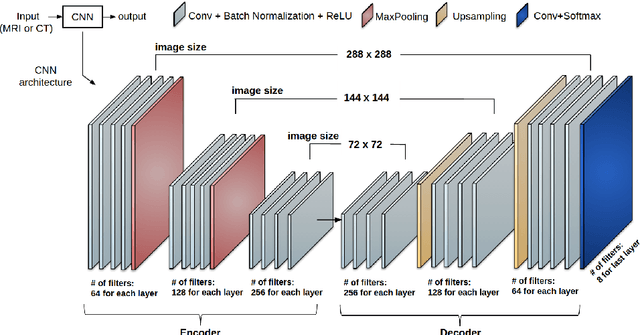
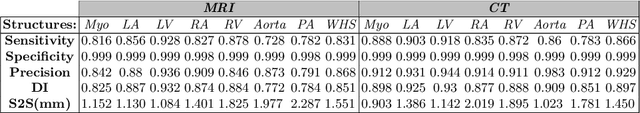
Abstract:Non-invasive detection of cardiovascular disorders from radiology scans requires quantitative image analysis of the heart and its substructures. There are well-established measurements that radiologists use for diseases assessment such as ejection fraction, volume of four chambers, and myocardium mass. These measurements are derived as outcomes of precise segmentation of the heart and its substructures. The aim of this paper is to provide such measurements through an accurate image segmentation algorithm that automatically delineates seven substructures of the heart from MRI and/or CT scans. Our proposed method is based on multi-planar deep convolutional neural networks (CNN) with an adaptive fusion strategy where we automatically utilize complementary information from different planes of the 3D scans for improved delineations. For CT and MRI, we have separately designed three CNNs (the same architectural configuration) for three planes, and have trained the networks from scratch for voxel-wise labeling for the following cardiac structures: myocardium of left ventricle (Myo), left atrium (LA), left ventricle (LV), right atrium (RA), right ventricle (RV), ascending aorta (Ao), and main pulmonary artery (PA). We have evaluated the proposed method with 4-fold-cross validation on the multi-modality whole heart segmentation challenge (MM-WHS 2017) dataset. The precision and dice index of 0.93 and 0.90, and 0.87 and 0.85 were achieved for CT and MR images, respectively. While a CT volume was segmented about 50 seconds, an MRI scan was segmented around 17 seconds with the GPUs/CUDA implementation.
CardiacNET: Segmentation of Left Atrium and Proximal Pulmonary Veins from MRI Using Multi-View CNN
May 19, 2017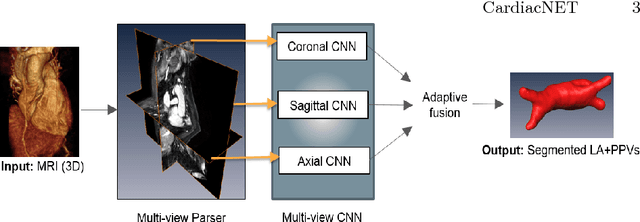
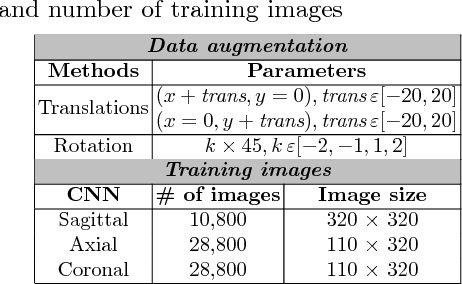
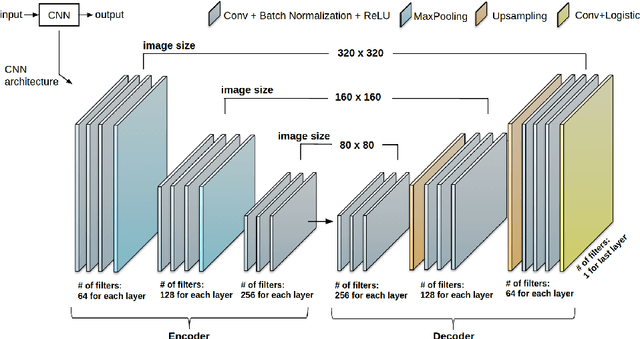
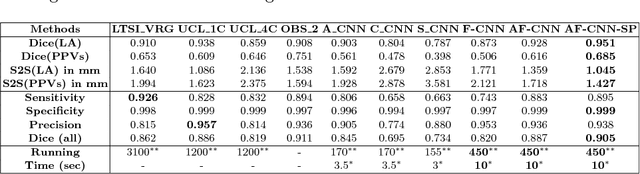
Abstract:Anatomical and biophysical modeling of left atrium (LA) and proximal pulmonary veins (PPVs) is important for clinical management of several cardiac diseases. Magnetic resonance imaging (MRI) allows qualitative assessment of LA and PPVs through visualization. However, there is a strong need for an advanced image segmentation method to be applied to cardiac MRI for quantitative analysis of LA and PPVs. In this study, we address this unmet clinical need by exploring a new deep learning-based segmentation strategy for quantification of LA and PPVs with high accuracy and heightened efficiency. Our approach is based on a multi-view convolutional neural network (CNN) with an adaptive fusion strategy and a new loss function that allows fast and more accurate convergence of the backpropagation based optimization. After training our network from scratch by using more than 60K 2D MRI images (slices), we have evaluated our segmentation strategy to the STACOM 2013 cardiac segmentation challenge benchmark. Qualitative and quantitative evaluations, obtained from the segmentation challenge, indicate that the proposed method achieved the state-of-the-art sensitivity (90%), specificity (99%), precision (94%), and efficiency levels (10 seconds in GPU, and 7.5 minutes in CPU).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge