Asma Saadia
Towards Low-Cost and Efficient Malaria Detection
Nov 26, 2021



Abstract:Malaria, a fatal but curable disease claims hundreds of thousands of lives every year. Early and correct diagnosis is vital to avoid health complexities, however, it depends upon the availability of costly microscopes and trained experts to analyze blood-smear slides. Deep learning-based methods have the potential to not only decrease the burden of experts but also improve diagnostic accuracy on low-cost microscopes. However, this is hampered by the absence of a reasonable size dataset. One of the most challenging aspects is the reluctance of the experts to annotate the dataset at low magnification on low-cost microscopes. We present a dataset to further the research on malaria microscopy over the low-cost microscopes at low magnification. Our large-scale dataset consists of images of blood-smear slides from several malaria-infected patients, collected through microscopes at two different cost spectrums and multiple magnifications. Malarial cells are annotated for the localization and life-stage classification task on the images collected through the high-cost microscope at high magnification. We design a mechanism to transfer these annotations from the high-cost microscope at high magnification to the low-cost microscope, at multiple magnifications. Multiple object detectors and domain adaptation methods are presented as the baselines. Furthermore, a partially supervised domain adaptation method is introduced to adapt the object-detector to work on the images collected from the low-cost microscope. The dataset will be made publicly available after publication.
Out of distribution detection for skin and malaria images
Nov 02, 2021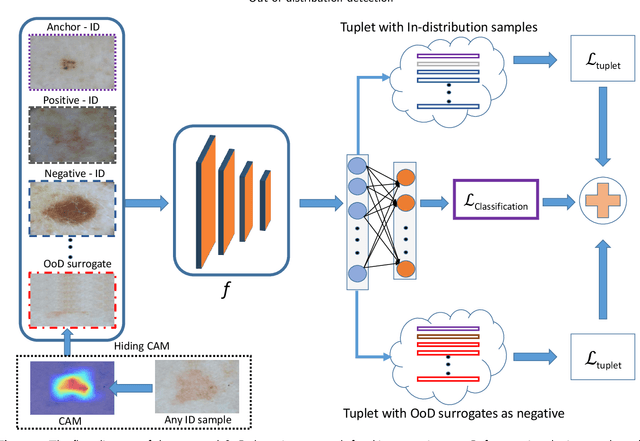
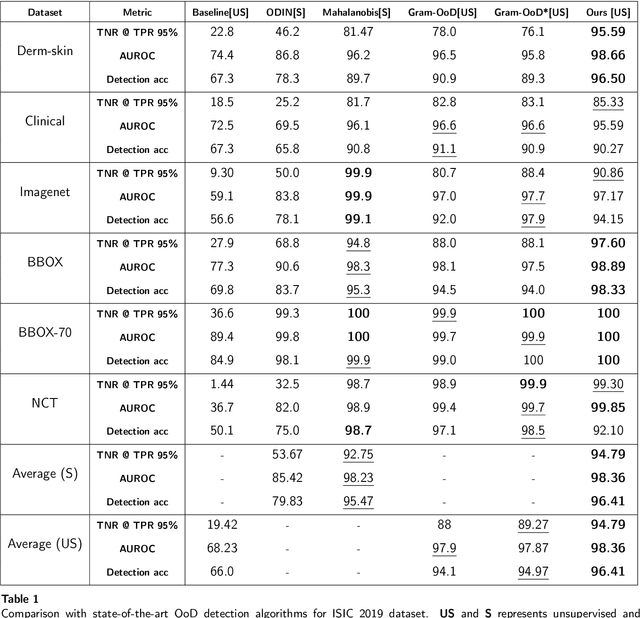
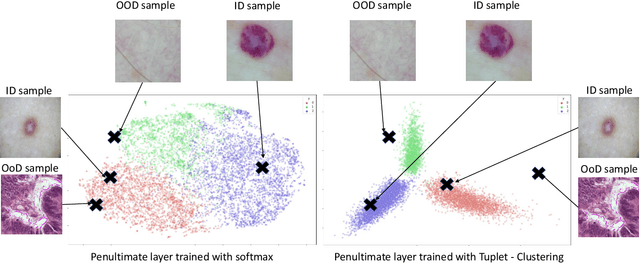
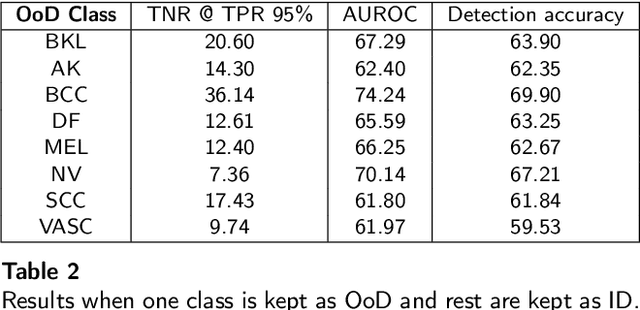
Abstract:Deep neural networks have shown promising results in disease detection and classification using medical image data. However, they still suffer from the challenges of handling real-world scenarios especially reliably detecting out-of-distribution (OoD) samples. We propose an approach to robustly classify OoD samples in skin and malaria images without the need to access labeled OoD samples during training. Specifically, we use metric learning along with logistic regression to force the deep networks to learn much rich class representative features. To guide the learning process against the OoD examples, we generate ID similar-looking examples by either removing class-specific salient regions in the image or permuting image parts and distancing them away from in-distribution samples. During inference time, the K-reciprocal nearest neighbor is employed to detect out-of-distribution samples. For skin cancer OoD detection, we employ two standard benchmark skin cancer ISIC datasets as ID, and six different datasets with varying difficulty levels were taken as out of distribution. For malaria OoD detection, we use the BBBC041 malaria dataset as ID and five different challenging datasets as out of distribution. We achieved state-of-the-art results, improving 5% and 4% in TNR@TPR95% over the previous state-of-the-art for skin cancer and malaria OoD detection respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge