Syed Javed
Region Guided Attention Network for Retinal Vessel Segmentation
Jul 22, 2024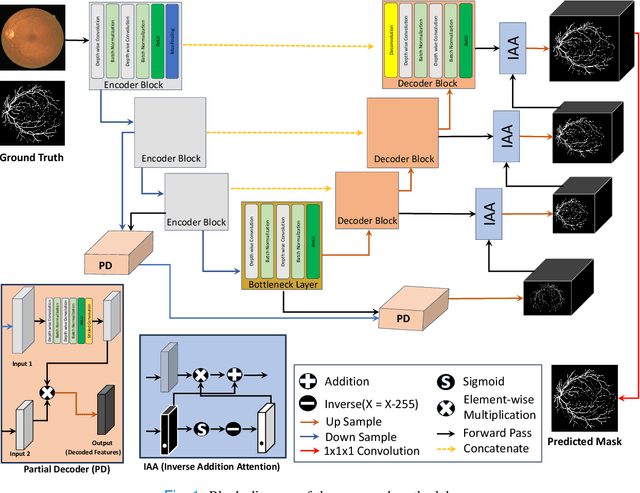
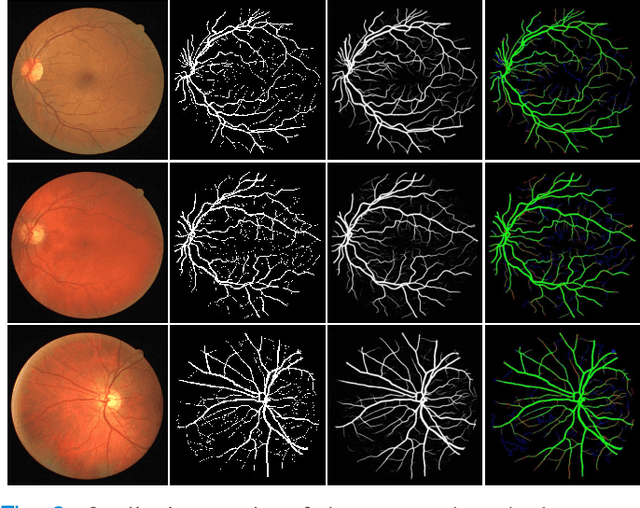
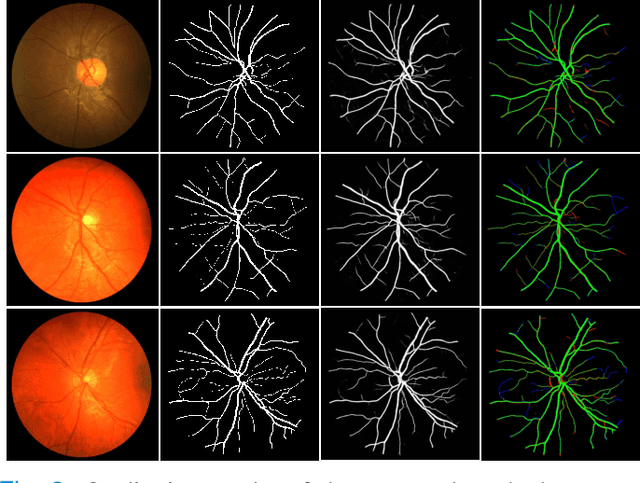
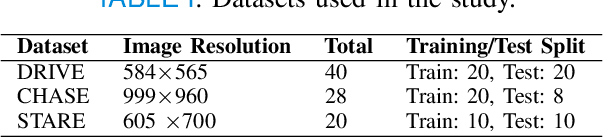
Abstract:Retinal imaging has emerged as a promising method of addressing this challenge, taking advantage of the unique structure of the retina. The retina is an embryonic extension of the central nervous system, providing a direct in vivo window into neurological health. Recent studies have shown that specific structural changes in retinal vessels can not only serve as early indicators of various diseases but also help to understand disease progression. In this work, we present a lightweight retinal vessel segmentation network based on the encoder-decoder mechanism with region-guided attention. We introduce inverse addition attention blocks with region guided attention to focus on the foreground regions and improve the segmentation of regions of interest. To further boost the model's performance on retinal vessel segmentation, we employ a weighted dice loss. This choice is particularly effective in addressing the class imbalance issues frequently encountered in retinal vessel segmentation tasks. Dice loss penalises false positives and false negatives equally, encouraging the model to generate more accurate segmentation with improved object boundary delineation and reduced fragmentation. Extensive experiments on a benchmark dataset show better performance (0.8285, 0.8098, 0.9677, and 0.8166 recall, precision, accuracy and F1 score respectively) compared to state-of-the-art methods.
Advancing Medical Image Segmentation with Mini-Net: A Lightweight Solution Tailored for Efficient Segmentation of Medical Images
May 27, 2024Abstract:Accurate segmentation of anatomical structures and abnormalities in medical images is crucial for computer-aided diagnosis and analysis. While deep learning techniques excel at this task, their computational demands pose challenges. Additionally, some cutting-edge segmentation methods, though effective for general object segmentation, may not be optimised for medical images. To address these issues, we propose Mini-Net, a lightweight segmentation network specifically designed for medical images. With fewer than 38,000 parameters, Mini-Net efficiently captures both high- and low-frequency features, enabling real-time applications in various medical imaging scenarios. We evaluate Mini-Net on various datasets, including DRIVE, STARE, ISIC-2016, ISIC-2018, and MoNuSeg, demonstrating its robustness and good performance compared to state-of-the-art methods.
Towards Low-Cost and Efficient Malaria Detection
Nov 26, 2021



Abstract:Malaria, a fatal but curable disease claims hundreds of thousands of lives every year. Early and correct diagnosis is vital to avoid health complexities, however, it depends upon the availability of costly microscopes and trained experts to analyze blood-smear slides. Deep learning-based methods have the potential to not only decrease the burden of experts but also improve diagnostic accuracy on low-cost microscopes. However, this is hampered by the absence of a reasonable size dataset. One of the most challenging aspects is the reluctance of the experts to annotate the dataset at low magnification on low-cost microscopes. We present a dataset to further the research on malaria microscopy over the low-cost microscopes at low magnification. Our large-scale dataset consists of images of blood-smear slides from several malaria-infected patients, collected through microscopes at two different cost spectrums and multiple magnifications. Malarial cells are annotated for the localization and life-stage classification task on the images collected through the high-cost microscope at high magnification. We design a mechanism to transfer these annotations from the high-cost microscope at high magnification to the low-cost microscope, at multiple magnifications. Multiple object detectors and domain adaptation methods are presented as the baselines. Furthermore, a partially supervised domain adaptation method is introduced to adapt the object-detector to work on the images collected from the low-cost microscope. The dataset will be made publicly available after publication.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge