Tariq M. Khan
Biological Brain Age Estimation using Sex-Aware Adversarial Variational Autoencoder with Multimodal Neuroimages
Dec 07, 2024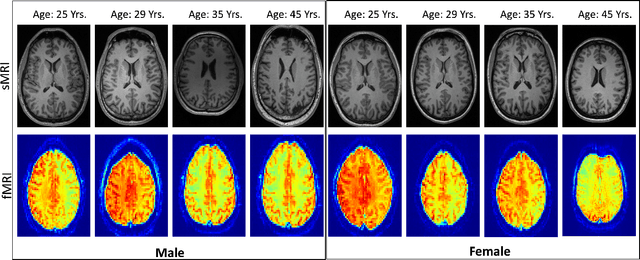
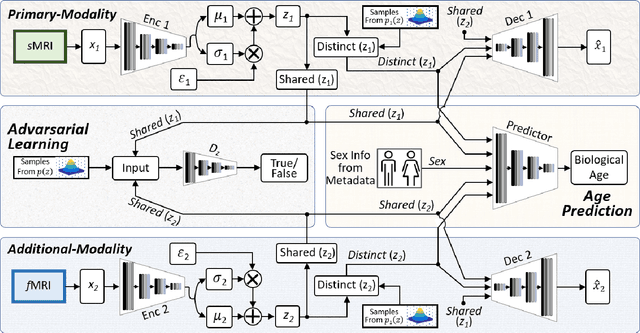
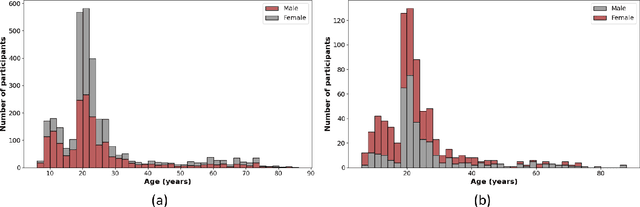
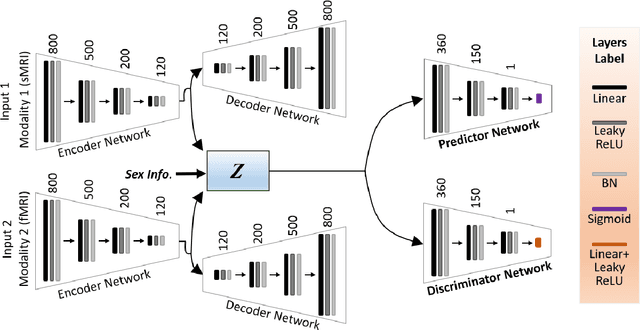
Abstract:Brain aging involves structural and functional changes and therefore serves as a key biomarker for brain health. Combining structural magnetic resonance imaging (sMRI) and functional magnetic resonance imaging (fMRI) has the potential to improve brain age estimation by leveraging complementary data. However, fMRI data, being noisier than sMRI, complicates multimodal fusion. Traditional fusion methods often introduce more noise than useful information, which can reduce accuracy compared to using sMRI alone. In this paper, we propose a novel multimodal framework for biological brain age estimation, utilizing a sex-aware adversarial variational autoencoder (SA-AVAE). Our framework integrates adversarial and variational learning to effectively disentangle the latent features from both modalities. Specifically, we decompose the latent space into modality-specific codes and shared codes to represent complementary and common information across modalities, respectively. To enhance the disentanglement, we introduce cross-reconstruction and shared-distinct distance ratio loss as regularization terms. Importantly, we incorporate sex information into the learned latent code, enabling the model to capture sex-specific aging patterns for brain age estimation via an integrated regressor module. We evaluate our model using the publicly available OpenBHB dataset, a comprehensive multi-site dataset for brain age estimation. The results from ablation studies and comparisons with state-of-the-art methods demonstrate that our framework outperforms existing approaches and shows significant robustness across various age groups, highlighting its potential for real-time clinical applications in the early detection of neurodegenerative diseases.
Multi-Task Adversarial Variational Autoencoder for Estimating Biological Brain Age with Multimodal Neuroimaging
Nov 15, 2024



Abstract:Despite advances in deep learning for estimating brain age from structural MRI data, incorporating functional MRI data is challenging due to its complex structure and the noisy nature of functional connectivity measurements. To address this, we present the Multitask Adversarial Variational Autoencoder, a custom deep learning framework designed to improve brain age predictions through multimodal MRI data integration. This model separates latent variables into generic and unique codes, isolating shared and modality-specific features. By integrating multitask learning with sex classification as an additional task, the model captures sex-specific aging patterns. Evaluated on the OpenBHB dataset, a large multisite brain MRI collection, the model achieves a mean absolute error of 2.77 years, outperforming traditional methods. This success positions M-AVAE as a powerful tool for metaverse-based healthcare applications in brain age estimation.
AD-Net: Attention-based dilated convolutional residual network with guided decoder for robust skin lesion segmentation
Sep 09, 2024Abstract:In computer-aided diagnosis tools employed for skin cancer treatment and early diagnosis, skin lesion segmentation is important. However, achieving precise segmentation is challenging due to inherent variations in appearance, contrast, texture, and blurry lesion boundaries. This research presents a robust approach utilizing a dilated convolutional residual network, which incorporates an attention-based spatial feature enhancement block (ASFEB) and employs a guided decoder strategy. In each dilated convolutional residual block, dilated convolution is employed to broaden the receptive field with varying dilation rates. To improve the spatial feature information of the encoder, we employed an attention-based spatial feature enhancement block in the skip connections. The ASFEB in our proposed method combines feature maps obtained from average and maximum-pooling operations. These combined features are then weighted using the active outcome of global average pooling and convolution operations. Additionally, we have incorporated a guided decoder strategy, where each decoder block is optimized using an individual loss function to enhance the feature learning process in the proposed AD-Net. The proposed AD-Net presents a significant benefit by necessitating fewer model parameters compared to its peer methods. This reduction in parameters directly impacts the number of labeled data required for training, facilitating faster convergence during the training process. The effectiveness of the proposed AD-Net was evaluated using four public benchmark datasets. We conducted a Wilcoxon signed-rank test to verify the efficiency of the AD-Net. The outcomes suggest that our method surpasses other cutting-edge methods in performance, even without the implementation of data augmentation strategies.
TBConvL-Net: A Hybrid Deep Learning Architecture for Robust Medical Image Segmentation
Sep 05, 2024



Abstract:Deep learning has shown great potential for automated medical image segmentation to improve the precision and speed of disease diagnostics. However, the task presents significant difficulties due to variations in the scale, shape, texture, and contrast of the pathologies. Traditional convolutional neural network (CNN) models have certain limitations when it comes to effectively modelling multiscale context information and facilitating information interaction between skip connections across levels. To overcome these limitations, a novel deep learning architecture is introduced for medical image segmentation, taking advantage of CNNs and vision transformers. Our proposed model, named TBConvL-Net, involves a hybrid network that combines the local features of a CNN encoder-decoder architecture with long-range and temporal dependencies using biconvolutional long-short-term memory (LSTM) networks and vision transformers (ViT). This enables the model to capture contextual channel relationships in the data and account for the uncertainty of segmentation over time. Additionally, we introduce a novel composite loss function that considers both the segmentation robustness and the boundary agreement of the predicted output with the gold standard. Our proposed model shows consistent improvement over the state of the art on ten publicly available datasets of seven different medical imaging modalities.
EUIS-Net: A Convolutional Neural Network for Efficient Ultrasound Image Segmentation
Aug 22, 2024



Abstract:Segmenting ultrasound images is critical for various medical applications, but it offers significant challenges due to ultrasound images' inherent noise and unpredictability. To address these challenges, we proposed EUIS-Net, a CNN network designed to segment ultrasound images efficiently and precisely. The proposed EUIS-Net utilises four encoder-decoder blocks, resulting in a notable decrease in computational complexity while achieving excellent performance. The proposed EUIS-Net integrates both channel and spatial attention mechanisms into the bottleneck to improve feature representation and collect significant contextual information. In addition, EUIS-Net incorporates a region-aware attention module in skip connections, which enhances the ability to concentrate on the region of the injury. To enable thorough information exchange across various network blocks, skip connection aggregation is employed from the network's lowermost to the uppermost block. Comprehensive evaluations are conducted on two publicly available ultrasound image segmentation datasets. The proposed EUIS-Net achieved mean IoU and dice scores of 78. 12\%, 85. 42\% and 84. 73\%, 89. 01\% in the BUSI and DDTI datasets, respectively. The findings of our study showcase the substantial capabilities of EUIS-Net for immediate use in clinical settings and its versatility in various ultrasound imaging tasks.
TESL-Net: A Transformer-Enhanced CNN for Accurate Skin Lesion Segmentation
Aug 19, 2024



Abstract:Early detection of skin cancer relies on precise segmentation of dermoscopic images of skin lesions. However, this task is challenging due to the irregular shape of the lesion, the lack of sharp borders, and the presence of artefacts such as marker colours and hair follicles. Recent methods for melanoma segmentation are U-Nets and fully connected networks (FCNs). As the depth of these neural network models increases, they can face issues like the vanishing gradient problem and parameter redundancy, potentially leading to a decrease in the Jaccard index of the segmentation model. In this study, we introduced a novel network named TESL-Net for the segmentation of skin lesions. The proposed TESL-Net involves a hybrid network that combines the local features of a CNN encoder-decoder architecture with long-range and temporal dependencies using bi-convolutional long-short-term memory (Bi-ConvLSTM) networks and a Swin transformer. This enables the model to account for the uncertainty of segmentation over time and capture contextual channel relationships in the data. We evaluated the efficacy of TESL-Net in three commonly used datasets (ISIC 2016, ISIC 2017, and ISIC 2018) for the segmentation of skin lesions. The proposed TESL-Net achieves state-of-the-art performance, as evidenced by a significantly elevated Jaccard index demonstrated by empirical results.
A Robust Algorithm for Contactless Fingerprint Enhancement and Matching
Aug 18, 2024



Abstract:Compared to contact fingerprint images, contactless fingerprint images exhibit four distinct characteristics: (1) they contain less noise; (2) they have fewer discontinuities in ridge patterns; (3) the ridge-valley pattern is less distinct; and (4) they pose an interoperability problem, as they lack the elastic deformation caused by pressing the finger against the capture device. These properties present significant challenges for the enhancement of contactless fingerprint images. In this study, we propose a novel contactless fingerprint identification solution that enhances the accuracy of minutiae detection through improved frequency estimation and a new region-quality-based minutia extraction algorithm. In addition, we introduce an efficient and highly accurate minutiae-based encoding and matching algorithm. We validate the effectiveness of our approach through extensive experimental testing. Our method achieves a minimum Equal Error Rate (EER) of 2.84\% on the PolyU contactless fingerprint dataset, demonstrating its superior performance compared to existing state-of-the-art techniques. The proposed fingerprint identification method exhibits notable precision and resilience, proving to be an effective and feasible solution for contactless fingerprint-based identification systems.
Region Guided Attention Network for Retinal Vessel Segmentation
Jul 22, 2024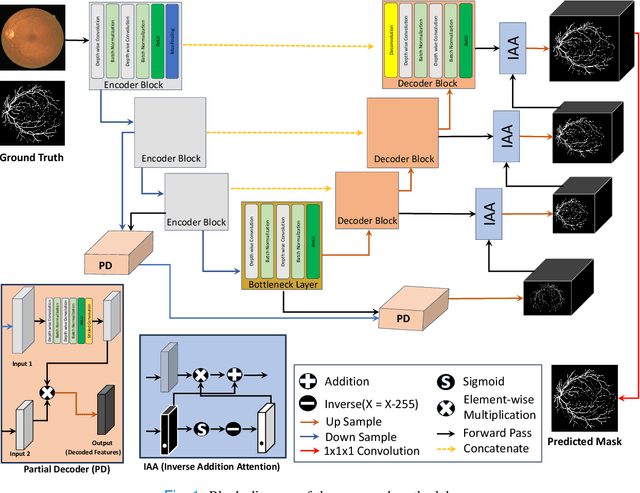
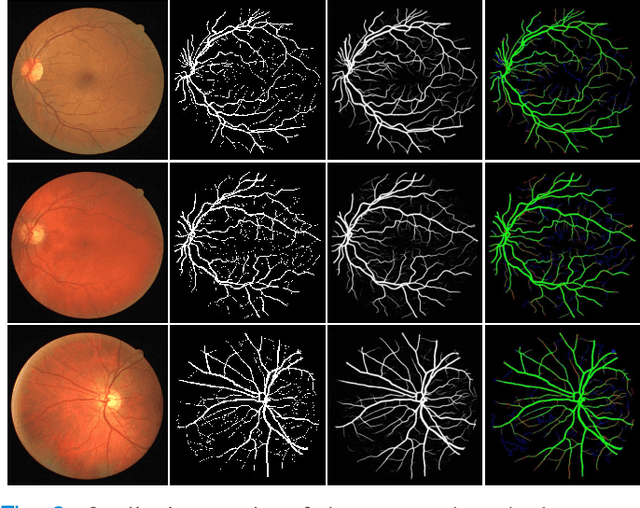
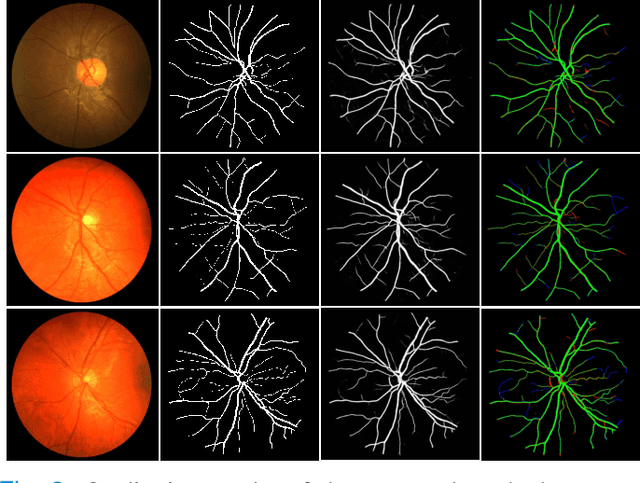
Abstract:Retinal imaging has emerged as a promising method of addressing this challenge, taking advantage of the unique structure of the retina. The retina is an embryonic extension of the central nervous system, providing a direct in vivo window into neurological health. Recent studies have shown that specific structural changes in retinal vessels can not only serve as early indicators of various diseases but also help to understand disease progression. In this work, we present a lightweight retinal vessel segmentation network based on the encoder-decoder mechanism with region-guided attention. We introduce inverse addition attention blocks with region guided attention to focus on the foreground regions and improve the segmentation of regions of interest. To further boost the model's performance on retinal vessel segmentation, we employ a weighted dice loss. This choice is particularly effective in addressing the class imbalance issues frequently encountered in retinal vessel segmentation tasks. Dice loss penalises false positives and false negatives equally, encouraging the model to generate more accurate segmentation with improved object boundary delineation and reduced fragmentation. Extensive experiments on a benchmark dataset show better performance (0.8285, 0.8098, 0.9677, and 0.8166 recall, precision, accuracy and F1 score respectively) compared to state-of-the-art methods.
Advancing Medical Image Segmentation with Mini-Net: A Lightweight Solution Tailored for Efficient Segmentation of Medical Images
May 27, 2024Abstract:Accurate segmentation of anatomical structures and abnormalities in medical images is crucial for computer-aided diagnosis and analysis. While deep learning techniques excel at this task, their computational demands pose challenges. Additionally, some cutting-edge segmentation methods, though effective for general object segmentation, may not be optimised for medical images. To address these issues, we propose Mini-Net, a lightweight segmentation network specifically designed for medical images. With fewer than 38,000 parameters, Mini-Net efficiently captures both high- and low-frequency features, enabling real-time applications in various medical imaging scenarios. We evaluate Mini-Net on various datasets, including DRIVE, STARE, ISIC-2016, ISIC-2018, and MoNuSeg, demonstrating its robustness and good performance compared to state-of-the-art methods.
LMBiS-Net: A Lightweight Multipath Bidirectional Skip Connection based CNN for Retinal Blood Vessel Segmentation
Sep 10, 2023Abstract:Blinding eye diseases are often correlated with altered retinal morphology, which can be clinically identified by segmenting retinal structures in fundus images. However, current methodologies often fall short in accurately segmenting delicate vessels. Although deep learning has shown promise in medical image segmentation, its reliance on repeated convolution and pooling operations can hinder the representation of edge information, ultimately limiting overall segmentation accuracy. In this paper, we propose a lightweight pixel-level CNN named LMBiS-Net for the segmentation of retinal vessels with an exceptionally low number of learnable parameters \textbf{(only 0.172 M)}. The network used multipath feature extraction blocks and incorporates bidirectional skip connections for the information flow between the encoder and decoder. Additionally, we have optimized the efficiency of the model by carefully selecting the number of filters to avoid filter overlap. This optimization significantly reduces training time and enhances computational efficiency. To assess the robustness and generalizability of LMBiS-Net, we performed comprehensive evaluations on various aspects of retinal images. Specifically, the model was subjected to rigorous tests to accurately segment retinal vessels, which play a vital role in ophthalmological diagnosis and treatment. By focusing on the retinal blood vessels, we were able to thoroughly analyze the performance and effectiveness of the LMBiS-Net model. The results of our tests demonstrate that LMBiS-Net is not only robust and generalizable but also capable of maintaining high levels of segmentation accuracy. These characteristics highlight the potential of LMBiS-Net as an efficient tool for high-speed and accurate segmentation of retinal images in various clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge