Arcot Sowmya
for the Sydney Memory and Ageing Study and the Alzheimer's Disease Neuroimaging Initiative
YOLO11-4K: An Efficient Architecture for Real-Time Small Object Detection in 4K Panoramic Images
Dec 18, 2025Abstract:The processing of omnidirectional 360-degree images poses significant challenges for object detection due to inherent spatial distortions, wide fields of view, and ultra-high-resolution inputs. Conventional detectors such as YOLO are optimised for standard image sizes (for example, 640x640 pixels) and often struggle with the computational demands of 4K or higher-resolution imagery typical of 360-degree vision. To address these limitations, we introduce YOLO11-4K, an efficient real-time detection framework tailored for 4K panoramic images. The architecture incorporates a novel multi-scale detection head with a P2 layer to improve sensitivity to small objects often missed at coarser scales, and a GhostConv-based backbone to reduce computational complexity without sacrificing representational power. To enable evaluation, we manually annotated the CVIP360 dataset, generating 6,876 frame-level bounding boxes and producing a publicly available, detection-ready benchmark for 4K panoramic scenes. YOLO11-4K achieves 0.95 mAP at 0.50 IoU with 28.3 milliseconds inference per frame, representing a 75 percent latency reduction compared to YOLO11 (112.3 milliseconds), while also improving accuracy (mAP at 0.50 of 0.95 versus 0.908). This balance of efficiency and precision enables robust object detection in expansive 360-degree environments, making the framework suitable for real-world high-resolution panoramic applications. While this work focuses on 4K omnidirectional images, the approach is broadly applicable to high-resolution detection tasks in autonomous navigation, surveillance, and augmented reality.
Predicting Coronary Artery Calcium Severity based on Non-Contrast Cardiac CT images using Deep Learning
Nov 10, 2025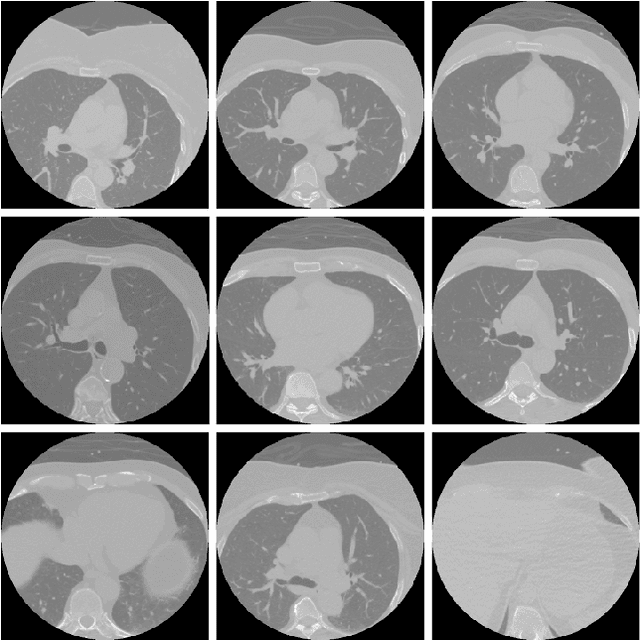
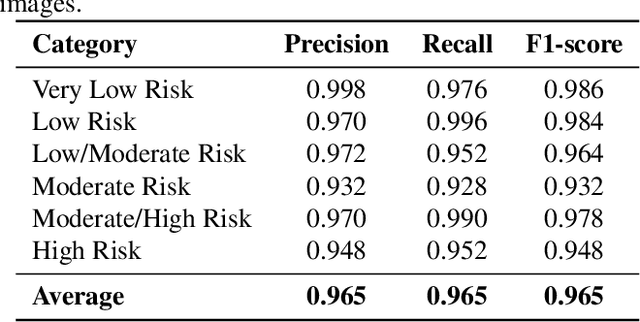
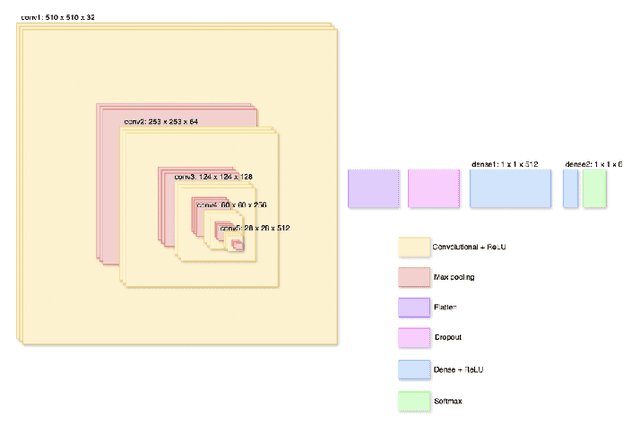
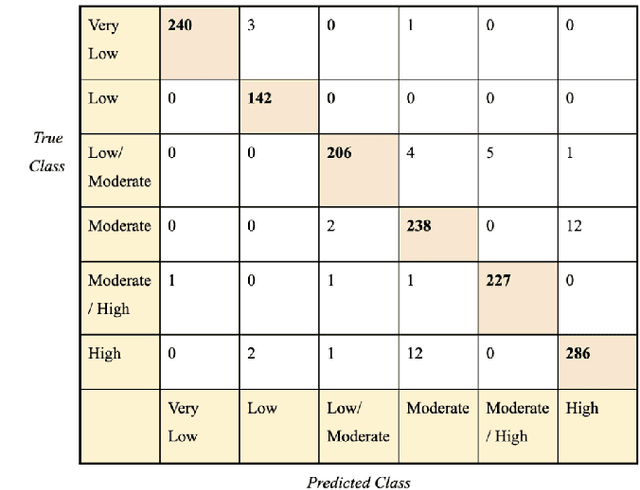
Abstract:Cardiovascular disease causes high rates of mortality worldwide. Coronary artery calcium (CAC) scoring is a powerful tool to stratify the risk of atherosclerotic cardiovascular disease. Current scoring practices require time-intensive semiautomatic analysis of cardiac computed tomography by radiologists and trained radiographers. The purpose of this study is to develop a deep learning convolutional neural networks (CNN) model to classify the calcium score in cardiac, non-contrast computed tomography images into one of six clinical categories. A total of 68 patient scans were retrospectively obtained together with their respective reported semiautomatic calcium score using an ECG-gated GE Discovery 570 Cardiac SPECT/CT camera. The dataset was divided into training, validation and test sets. Using the semiautomatic CAC score as the reference label, the model demonstrated high performance on a six-class CAC scoring categorisation task. Of the scans analysed, the model misclassified 32 cases, tending towards overestimating the CAC in 26 out of 32 misclassifications. Overall, the model showed high agreement (Cohen's kappa of 0.962), an overall accuracy of 96.5% and high generalisability. The results suggest that the model outputs were accurate and consistent with current semiautomatic practice, with good generalisability to test data. The model demonstrates the viability of a CNN model to stratify the calcium score into an expanded set of six clinical categories.
LWT-ARTERY-LABEL: A Lightweight Framework for Automated Coronary Artery Identification
Aug 09, 2025Abstract:Coronary artery disease (CAD) remains the leading cause of death globally, with computed tomography coronary angiography (CTCA) serving as a key diagnostic tool. However, coronary arterial analysis using CTCA, such as identifying artery-specific features from computational modelling, is labour-intensive and time-consuming. Automated anatomical labelling of coronary arteries offers a potential solution, yet the inherent anatomical variability of coronary trees presents a significant challenge. Traditional knowledge-based labelling methods fall short in leveraging data-driven insights, while recent deep-learning approaches often demand substantial computational resources and overlook critical clinical knowledge. To address these limitations, we propose a lightweight method that integrates anatomical knowledge with rule-based topology constraints for effective coronary artery labelling. Our approach achieves state-of-the-art performance on benchmark datasets, providing a promising alternative for automated coronary artery labelling.
GRAPHITE: Graph-Based Interpretable Tissue Examination for Enhanced Explainability in Breast Cancer Histopathology
Jan 08, 2025



Abstract:Explainable AI (XAI) in medical histopathology is essential for enhancing the interpretability and clinical trustworthiness of deep learning models in cancer diagnosis. However, the black-box nature of these models often limits their clinical adoption. We introduce GRAPHITE (Graph-based Interpretable Tissue Examination), a post-hoc explainable framework designed for breast cancer tissue microarray (TMA) analysis. GRAPHITE employs a multiscale approach, extracting patches at various magnification levels, constructing an hierarchical graph, and utilising graph attention networks (GAT) with scalewise attention (SAN) to capture scale-dependent features. We trained the model on 140 tumour TMA cores and four benign whole slide images from which 140 benign samples were created, and tested it on 53 pathologist-annotated TMA samples. GRAPHITE outperformed traditional XAI methods, achieving a mean average precision (mAP) of 0.56, an area under the receiver operating characteristic curve (AUROC) of 0.94, and a threshold robustness (ThR) of 0.70, indicating that the model maintains high performance across a wide range of thresholds. In clinical utility, GRAPHITE achieved the highest area under the decision curve (AUDC) of 4.17e+5, indicating reliable decision support across thresholds. These results highlight GRAPHITE's potential as a clinically valuable tool in computational pathology, providing interpretable visualisations that align with the pathologists' diagnostic reasoning and support precision medicine.
Region Guided Attention Network for Retinal Vessel Segmentation
Jul 22, 2024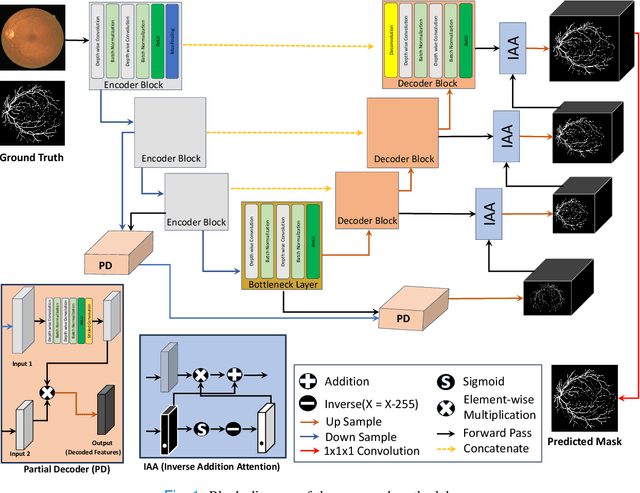
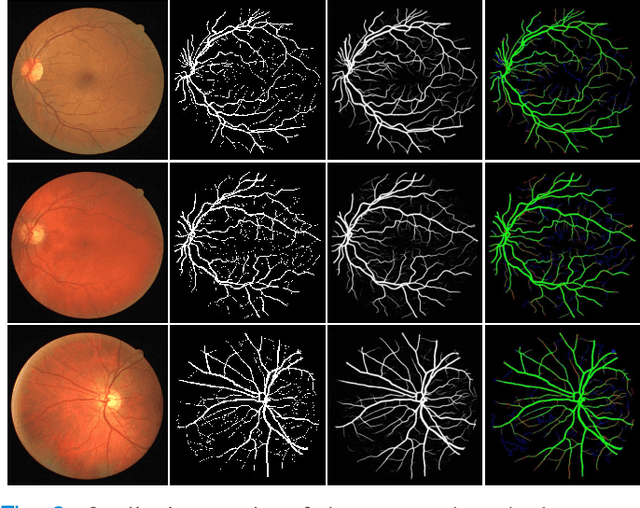
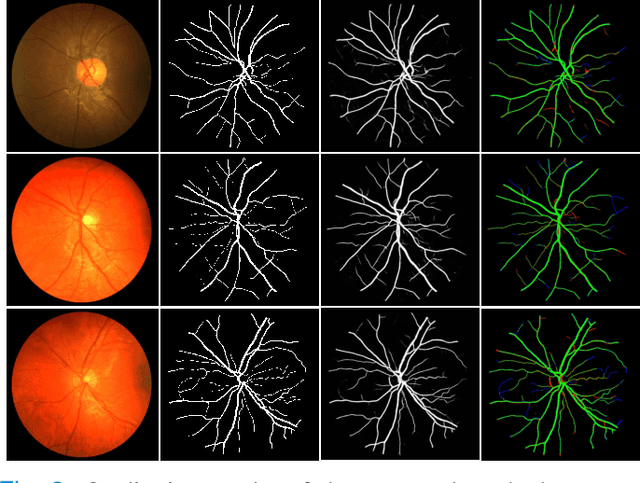
Abstract:Retinal imaging has emerged as a promising method of addressing this challenge, taking advantage of the unique structure of the retina. The retina is an embryonic extension of the central nervous system, providing a direct in vivo window into neurological health. Recent studies have shown that specific structural changes in retinal vessels can not only serve as early indicators of various diseases but also help to understand disease progression. In this work, we present a lightweight retinal vessel segmentation network based on the encoder-decoder mechanism with region-guided attention. We introduce inverse addition attention blocks with region guided attention to focus on the foreground regions and improve the segmentation of regions of interest. To further boost the model's performance on retinal vessel segmentation, we employ a weighted dice loss. This choice is particularly effective in addressing the class imbalance issues frequently encountered in retinal vessel segmentation tasks. Dice loss penalises false positives and false negatives equally, encouraging the model to generate more accurate segmentation with improved object boundary delineation and reduced fragmentation. Extensive experiments on a benchmark dataset show better performance (0.8285, 0.8098, 0.9677, and 0.8166 recall, precision, accuracy and F1 score respectively) compared to state-of-the-art methods.
Advancing Medical Image Segmentation with Mini-Net: A Lightweight Solution Tailored for Efficient Segmentation of Medical Images
May 27, 2024Abstract:Accurate segmentation of anatomical structures and abnormalities in medical images is crucial for computer-aided diagnosis and analysis. While deep learning techniques excel at this task, their computational demands pose challenges. Additionally, some cutting-edge segmentation methods, though effective for general object segmentation, may not be optimised for medical images. To address these issues, we propose Mini-Net, a lightweight segmentation network specifically designed for medical images. With fewer than 38,000 parameters, Mini-Net efficiently captures both high- and low-frequency features, enabling real-time applications in various medical imaging scenarios. We evaluate Mini-Net on various datasets, including DRIVE, STARE, ISIC-2016, ISIC-2018, and MoNuSeg, demonstrating its robustness and good performance compared to state-of-the-art methods.
MM-SurvNet: Deep Learning-Based Survival Risk Stratification in Breast Cancer Through Multimodal Data Fusion
Feb 19, 2024Abstract:Survival risk stratification is an important step in clinical decision making for breast cancer management. We propose a novel deep learning approach for this purpose by integrating histopathological imaging, genetic and clinical data. It employs vision transformers, specifically the MaxViT model, for image feature extraction, and self-attention to capture intricate image relationships at the patient level. A dual cross-attention mechanism fuses these features with genetic data, while clinical data is incorporated at the final layer to enhance predictive accuracy. Experiments on the public TCGA-BRCA dataset show that our model, trained using the negative log likelihood loss function, can achieve superior performance with a mean C-index of 0.64, surpassing existing methods. This advancement facilitates tailored treatment strategies, potentially leading to improved patient outcomes.
BioFusionNet: Deep Learning-Based Survival Risk Stratification in ER+ Breast Cancer Through Multifeature and Multimodal Data Fusion
Feb 16, 2024Abstract:Breast cancer is a significant health concern affecting millions of women worldwide. Accurate survival risk stratification plays a crucial role in guiding personalised treatment decisions and improving patient outcomes. Here we present BioFusionNet, a deep learning framework that fuses image-derived features with genetic and clinical data to achieve a holistic patient profile and perform survival risk stratification of ER+ breast cancer patients. We employ multiple self-supervised feature extractors, namely DINO and MoCoV3, pretrained on histopathology patches to capture detailed histopathological image features. We then utilise a variational autoencoder (VAE) to fuse these features, and harness the latent space of the VAE to feed into a self-attention network, generating patient-level features. Next, we develop a co-dual-cross-attention mechanism to combine the histopathological features with genetic data, enabling the model to capture the interplay between them. Additionally, clinical data is incorporated using a feed-forward network (FFN), further enhancing predictive performance and achieving comprehensive multimodal feature integration. Furthermore, we introduce a weighted Cox loss function, specifically designed to handle imbalanced survival data, which is a common challenge in the field. The proposed model achieves a mean concordance index (C-index) of 0.77 and a time-dependent area under the curve (AUC) of 0.84, outperforming state-of-the-art methods. It predicts risk (high versus low) with prognostic significance for overall survival (OS) in univariate analysis (HR=2.99, 95% CI: 1.88--4.78, p<0.005), and maintains independent significance in multivariate analysis incorporating standard clinicopathological variables (HR=2.91, 95% CI: 1.80--4.68, p<0.005). The proposed method not only improves model performance but also addresses a critical gap in handling imbalanced data.
Automatic 3D Multi-modal Ultrasound Segmentation of Human Placenta using Fusion Strategies and Deep Learning
Jan 17, 2024Abstract:Purpose: Ultrasound is the most commonly used medical imaging modality for diagnosis and screening in clinical practice. Due to its safety profile, noninvasive nature and portability, ultrasound is the primary imaging modality for fetal assessment in pregnancy. Current ultrasound processing methods are either manual or semi-automatic and are therefore laborious, time-consuming and prone to errors, and automation would go a long way in addressing these challenges. Automated identification of placental changes at earlier gestation could facilitate potential therapies for conditions such as fetal growth restriction and pre-eclampsia that are currently detected only at late gestational age, potentially preventing perinatal morbidity and mortality. Methods: We propose an automatic three-dimensional multi-modal (B-mode and power Doppler) ultrasound segmentation of the human placenta using deep learning combined with different fusion strategies.We collected data containing Bmode and power Doppler ultrasound scans for 400 studies. Results: We evaluated different fusion strategies and state-of-the-art image segmentation networks for placenta segmentation based on standard overlap- and boundary-based metrics. We found that multimodal information in the form of B-mode and power Doppler scans outperform any single modality. Furthermore, we found that B-mode and power Doppler input scans fused at the data level provide the best results with a mean Dice Similarity Coefficient (DSC) of 0.849. Conclusion: We conclude that the multi-modal approach of combining B-mode and power Doppler scans is effective in segmenting the placenta from 3D ultrasound scans in a fully automated manner and is robust to quality variation of the datasets.
Assessing Encoder-Decoder Architectures for Robust Coronary Artery Segmentation
Oct 16, 2023Abstract:Coronary artery diseases are among the leading causes of mortality worldwide. Timely and accurate diagnosis, facilitated by precise coronary artery segmentation, is pivotal in changing patient outcomes. In the realm of biomedical imaging, convolutional neural networks, especially the U-Net architecture, have revolutionised segmentation processes. However, one of the primary challenges remains the lack of benchmarking datasets specific to coronary arteries. However through the use of the recently published public dataset ASOCA, the potential of deep learning for accurate coronary segmentation can be improved. This paper delves deep into examining the performance of 25 distinct encoder-decoder combinations. Through analysis of the 40 cases provided to ASOCA participants, it is revealed that the EfficientNet-LinkNet combination, serving as encoder and decoder, stands out. It achieves a Dice coefficient of 0.882 and a 95th percentile Hausdorff distance of 4.753. These findings not only underscore the superiority of our model in comparison to those presented at the MICCAI 2020 challenge but also set the stage for future advancements in coronary artery segmentation, opening doors to enhanced diagnostic and treatment strategies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge