Azka Rehman
Dual-Encoder Transformer-Based Multimodal Learning for Ischemic Stroke Lesion Segmentation Using Diffusion MRI
Dec 23, 2025Abstract:Accurate segmentation of ischemic stroke lesions from diffusion magnetic resonance imaging (MRI) is essential for clinical decision-making and outcome assessment. Diffusion-Weighted Imaging (DWI) and Apparent Diffusion Coefficient (ADC) scans provide complementary information on acute and sub-acute ischemic changes; however, automated lesion delineation remains challenging due to variability in lesion appearance. In this work, we study ischemic stroke lesion segmentation using multimodal diffusion MRI from the ISLES 2022 dataset. Several state-of-the-art convolutional and transformer-based architectures, including U-Net variants, Swin-UNet, and TransUNet, are benchmarked. Based on performance, a dual-encoder TransUNet architecture is proposed to learn modality-specific representations from DWI and ADC inputs. To incorporate spatial context, adjacent slice information is integrated using a three-slice input configuration. All models are trained under a unified framework and evaluated using the Dice Similarity Coefficient (DSC). Results show that transformer-based models outperform convolutional baselines, and the proposed dual-encoder TransUNet achieves the best performance, reaching a Dice score of 85.4% on the test set. The proposed framework offers a robust solution for automated ischemic stroke lesion segmentation from diffusion MRI.
Anatomy-Guided Representation Learning Using a Transformer-Based Network for Thyroid Nodule Segmentation in Ultrasound Images
Dec 14, 2025Abstract:Accurate thyroid nodule segmentation in ultrasound images is critical for diagnosis and treatment planning. However, ambiguous boundaries between nodules and surrounding tissues, size variations, and the scarcity of annotated ultrasound data pose significant challenges for automated segmentation. Existing deep learning models struggle to incorporate contextual information from the thyroid gland and generalize effectively across diverse cases. To address these challenges, we propose SSMT-Net, a Semi-Supervised Multi-Task Transformer-based Network that leverages unlabeled data to enhance Transformer-centric encoder feature extraction capability in an initial unsupervised phase. In the supervised phase, the model jointly optimizes nodule segmentation, gland segmentation, and nodule size estimation, integrating both local and global contextual features. Extensive evaluations on the TN3K and DDTI datasets demonstrate that SSMT-Net outperforms state-of-the-art methods, with higher accuracy and robustness, indicating its potential for real-world clinical applications.
Biological Brain Age Estimation using Sex-Aware Adversarial Variational Autoencoder with Multimodal Neuroimages
Dec 07, 2024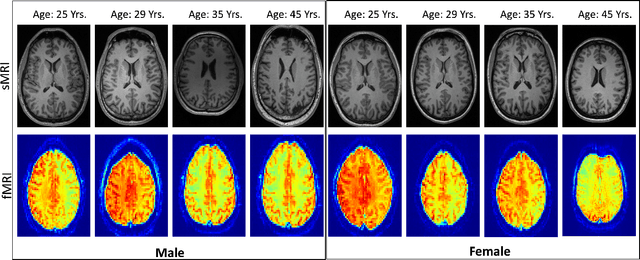
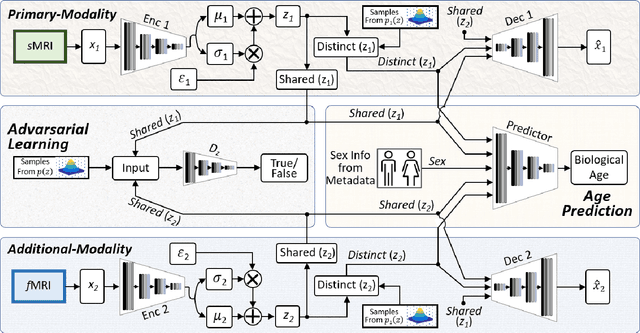
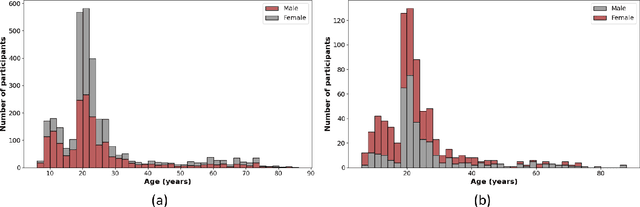
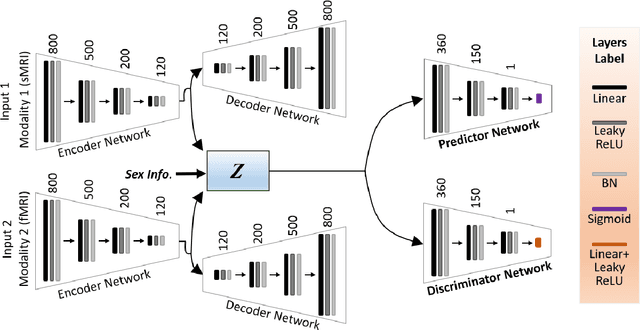
Abstract:Brain aging involves structural and functional changes and therefore serves as a key biomarker for brain health. Combining structural magnetic resonance imaging (sMRI) and functional magnetic resonance imaging (fMRI) has the potential to improve brain age estimation by leveraging complementary data. However, fMRI data, being noisier than sMRI, complicates multimodal fusion. Traditional fusion methods often introduce more noise than useful information, which can reduce accuracy compared to using sMRI alone. In this paper, we propose a novel multimodal framework for biological brain age estimation, utilizing a sex-aware adversarial variational autoencoder (SA-AVAE). Our framework integrates adversarial and variational learning to effectively disentangle the latent features from both modalities. Specifically, we decompose the latent space into modality-specific codes and shared codes to represent complementary and common information across modalities, respectively. To enhance the disentanglement, we introduce cross-reconstruction and shared-distinct distance ratio loss as regularization terms. Importantly, we incorporate sex information into the learned latent code, enabling the model to capture sex-specific aging patterns for brain age estimation via an integrated regressor module. We evaluate our model using the publicly available OpenBHB dataset, a comprehensive multi-site dataset for brain age estimation. The results from ablation studies and comparisons with state-of-the-art methods demonstrate that our framework outperforms existing approaches and shows significant robustness across various age groups, highlighting its potential for real-time clinical applications in the early detection of neurodegenerative diseases.
Multi-Task Adversarial Variational Autoencoder for Estimating Biological Brain Age with Multimodal Neuroimaging
Nov 15, 2024



Abstract:Despite advances in deep learning for estimating brain age from structural MRI data, incorporating functional MRI data is challenging due to its complex structure and the noisy nature of functional connectivity measurements. To address this, we present the Multitask Adversarial Variational Autoencoder, a custom deep learning framework designed to improve brain age predictions through multimodal MRI data integration. This model separates latent variables into generic and unique codes, isolating shared and modality-specific features. By integrating multitask learning with sex classification as an additional task, the model captures sex-specific aging patterns. Evaluated on the OpenBHB dataset, a large multisite brain MRI collection, the model achieves a mean absolute error of 2.77 years, outperforming traditional methods. This success positions M-AVAE as a powerful tool for metaverse-based healthcare applications in brain age estimation.
MESAHA-Net: Multi-Encoders based Self-Adaptive Hard Attention Network with Maximum Intensity Projections for Lung Nodule Segmentation in CT Scan
Apr 04, 2023Abstract:Accurate lung nodule segmentation is crucial for early-stage lung cancer diagnosis, as it can substantially enhance patient survival rates. Computed tomography (CT) images are widely employed for early diagnosis in lung nodule analysis. However, the heterogeneity of lung nodules, size diversity, and the complexity of the surrounding environment pose challenges for developing robust nodule segmentation methods. In this study, we propose an efficient end-to-end framework, the multi-encoder-based self-adaptive hard attention network (MESAHA-Net), for precise lung nodule segmentation in CT scans. MESAHA-Net comprises three encoding paths, an attention block, and a decoder block, facilitating the integration of three types of inputs: CT slice patches, forward and backward maximum intensity projection (MIP) images, and region of interest (ROI) masks encompassing the nodule. By employing a novel adaptive hard attention mechanism, MESAHA-Net iteratively performs slice-by-slice 2D segmentation of lung nodules, focusing on the nodule region in each slice to generate 3D volumetric segmentation of lung nodules. The proposed framework has been comprehensively evaluated on the LIDC-IDRI dataset, the largest publicly available dataset for lung nodule segmentation. The results demonstrate that our approach is highly robust for various lung nodule types, outperforming previous state-of-the-art techniques in terms of segmentation accuracy and computational complexity, rendering it suitable for real-time clinical implementation.
MEDS-Net: Self-Distilled Multi-Encoders Network with Bi-Direction Maximum Intensity projections for Lung Nodule Detection
Oct 30, 2022Abstract:In this study, we propose a lung nodule detection scheme which fully incorporates the clinic workflow of radiologists. Particularly, we exploit Bi-Directional Maximum intensity projection (MIP) images of various thicknesses (i.e., 3, 5 and 10mm) along with a 3D patch of CT scan, consisting of 10 adjacent slices to feed into self-distillation-based Multi-Encoders Network (MEDS-Net). The proposed architecture first condenses 3D patch input to three channels by using a dense block which consists of dense units which effectively examine the nodule presence from 2D axial slices. This condensed information, along with the forward and backward MIP images, is fed to three different encoders to learn the most meaningful representation, which is forwarded into the decoded block at various levels. At the decoder block, we employ a self-distillation mechanism by connecting the distillation block, which contains five lung nodule detectors. It helps to expedite the convergence and improves the learning ability of the proposed architecture. Finally, the proposed scheme reduces the false positives by complementing the main detector with auxiliary detectors. The proposed scheme has been rigorously evaluated on 888 scans of LUNA16 dataset and obtained a CPM score of 93.6\%. The results demonstrate that incorporating of bi-direction MIP images enables MEDS-Net to effectively distinguish nodules from surroundings which help to achieve the sensitivity of 91.5% and 92.8% with false positives rate of 0.25 and 0.5 per scan, respectively.
Dual-Stage Deeply Supervised Attention-based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans
Oct 06, 2022



Abstract:Accurate segmentation of mandibular canals in lower jaws is important in dental implantology, in which the implant position and dimensions are currently determined manually from 3D CT images by medical experts to avoid damaging the mandibular nerve inside the canal. In this paper, we propose a novel dual-stage deep learning based scheme for automatic detection of mandibular canal. Particularly, we first we enhance the CBCT scans by employing the novel histogram-based dynamic windowing scheme which improves the visibility of mandibular canals. After enhancement, we design 3D deeply supervised attention U-Net architecture for localize the volume of interest (VOI) which contains the mandibular canals (i.e., left and right canals). Finally, we employed the multi-scale input residual U-Net architecture (MS-R-UNet) to accurately segment the mandibular canals. The proposed method has been rigorously evaluated on 500 scans and results demonstrate that our technique out performs the existing state-of-the-art methods in term of segmentation performance as well as robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge