Pradip K. Das
Diverse Image Generation with Diffusion Models and Cross Class Label Learning for Polyp Classification
Feb 08, 2025



Abstract:Pathologic diagnosis is a critical phase in deciding the optimal treatment procedure for dealing with colorectal cancer (CRC). Colonic polyps, precursors to CRC, can pathologically be classified into two major types: adenomatous and hyperplastic. For precise classification and early diagnosis of such polyps, the medical procedure of colonoscopy has been widely adopted paired with various imaging techniques, including narrow band imaging and white light imaging. However, the existing classification techniques mainly rely on a single imaging modality and show limited performance due to data scarcity. Recently, generative artificial intelligence has been gaining prominence in overcoming such issues. Additionally, various generation-controlling mechanisms using text prompts and images have been introduced to obtain visually appealing and desired outcomes. However, such mechanisms require class labels to make the model respond efficiently to the provided control input. In the colonoscopy domain, such controlling mechanisms are rarely explored; specifically, the text prompt is a completely uninvestigated area. Moreover, the unavailability of expensive class-wise labels for diverse sets of images limits such explorations. Therefore, we develop a novel model, PathoPolyp-Diff, that generates text-controlled synthetic images with diverse characteristics in terms of pathology, imaging modalities, and quality. We introduce cross-class label learning to make the model learn features from other classes, reducing the burdensome task of data annotation. The experimental results report an improvement of up to 7.91% in balanced accuracy using a publicly available dataset. Moreover, cross-class label learning achieves a statistically significant improvement of up to 18.33% in balanced accuracy during video-level analysis. The code is available at https://github.com/Vanshali/PathoPolyp-Diff.
An objective validation of polyp and instrument segmentation methods in colonoscopy through Medico 2020 polyp segmentation and MedAI 2021 transparency challenges
Jul 30, 2023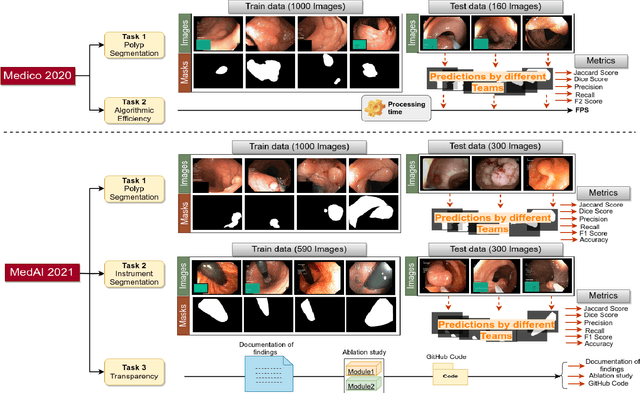
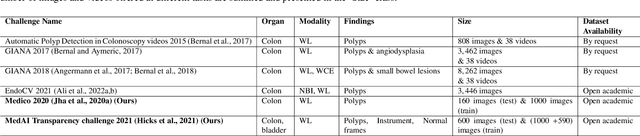
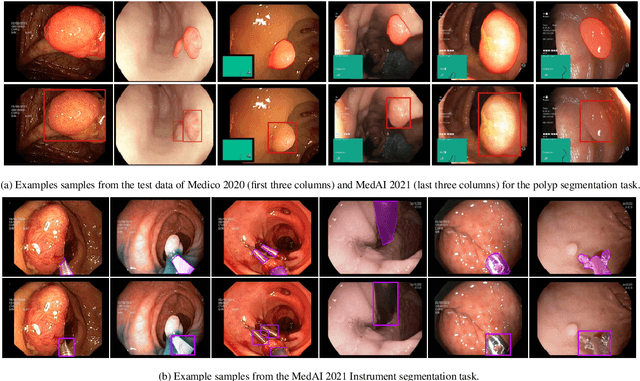
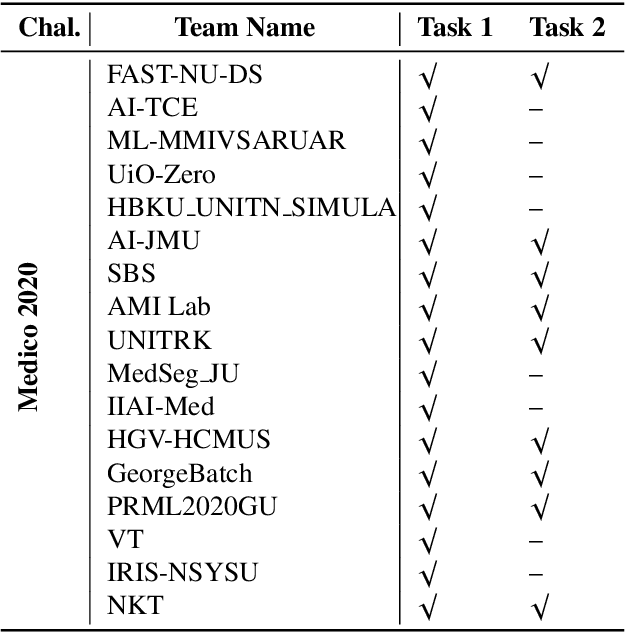
Abstract:Automatic analysis of colonoscopy images has been an active field of research motivated by the importance of early detection of precancerous polyps. However, detecting polyps during the live examination can be challenging due to various factors such as variation of skills and experience among the endoscopists, lack of attentiveness, and fatigue leading to a high polyp miss-rate. Deep learning has emerged as a promising solution to this challenge as it can assist endoscopists in detecting and classifying overlooked polyps and abnormalities in real time. In addition to the algorithm's accuracy, transparency and interpretability are crucial to explaining the whys and hows of the algorithm's prediction. Further, most algorithms are developed in private data, closed source, or proprietary software, and methods lack reproducibility. Therefore, to promote the development of efficient and transparent methods, we have organized the "Medico automatic polyp segmentation (Medico 2020)" and "MedAI: Transparency in Medical Image Segmentation (MedAI 2021)" competitions. We present a comprehensive summary and analyze each contribution, highlight the strength of the best-performing methods, and discuss the possibility of clinical translations of such methods into the clinic. For the transparency task, a multi-disciplinary team, including expert gastroenterologists, accessed each submission and evaluated the team based on open-source practices, failure case analysis, ablation studies, usability and understandability of evaluations to gain a deeper understanding of the models' credibility for clinical deployment. Through the comprehensive analysis of the challenge, we not only highlight the advancements in polyp and surgical instrument segmentation but also encourage qualitative evaluation for building more transparent and understandable AI-based colonoscopy systems.
GastroVision: A Multi-class Endoscopy Image Dataset for Computer Aided Gastrointestinal Disease Detection
Jul 16, 2023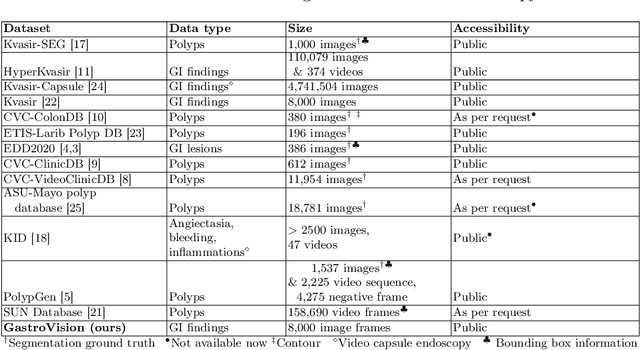
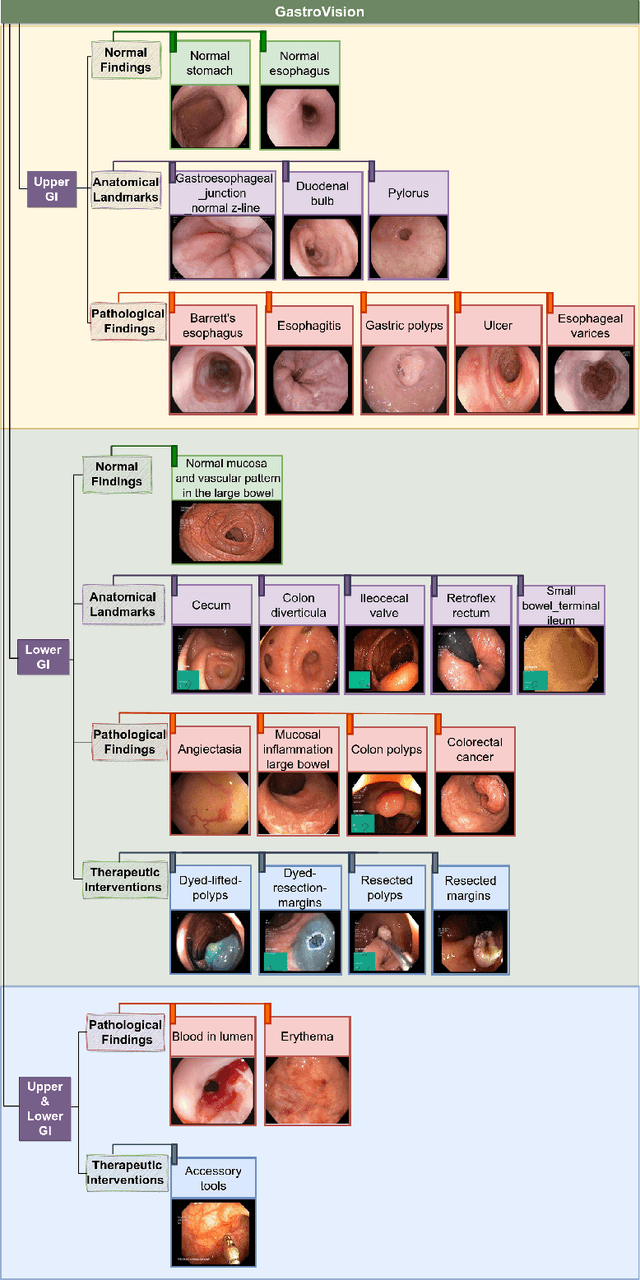
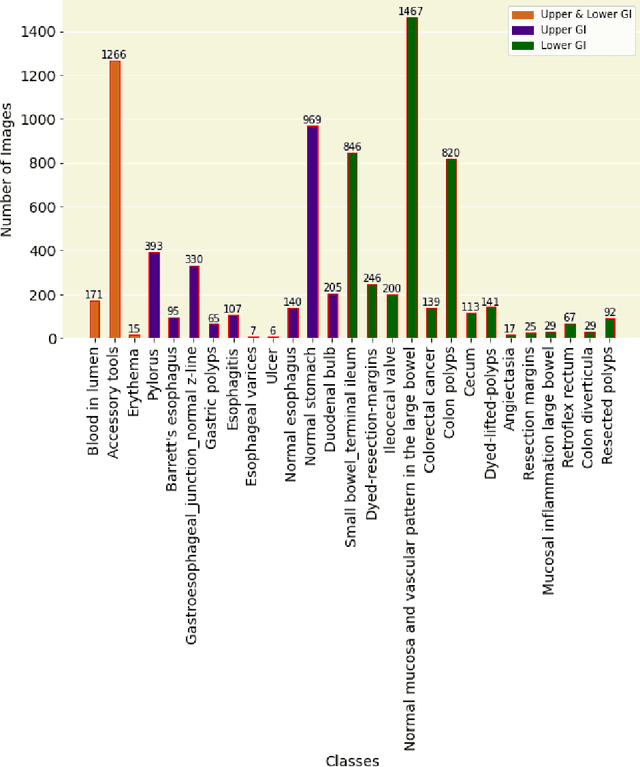
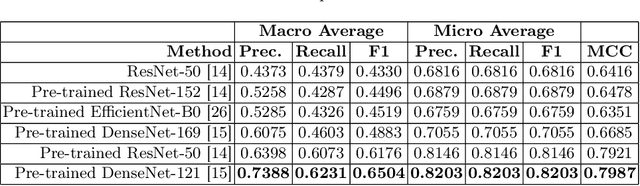
Abstract:Integrating real-time artificial intelligence (AI) systems in clinical practices faces challenges such as scalability and acceptance. These challenges include data availability, biased outcomes, data quality, lack of transparency, and underperformance on unseen datasets from different distributions. The scarcity of large-scale, precisely labeled, and diverse datasets are the major challenge for clinical integration. This scarcity is also due to the legal restrictions and extensive manual efforts required for accurate annotations from clinicians. To address these challenges, we present GastroVision, a multi-center open-access gastrointestinal (GI) endoscopy dataset that includes different anatomical landmarks, pathological abnormalities, polyp removal cases and normal findings (a total of 24 classes) from the GI tract. The dataset comprises 8,000 images acquired from B{\ae}rum Hospital in Norway and Karolinska University in Sweden and was annotated and verified by experienced GI endoscopists. Furthermore, we validate the significance of our dataset with extensive benchmarking based on the popular deep learning based baseline models. We believe our dataset can facilitate the development of AI-based algorithms for GI disease detection and classification. Our dataset is available at https://osf.io/84e7f/.
Can Adversarial Networks Make Uninformative Colonoscopy Video Frames Clinically Informative?
Apr 04, 2023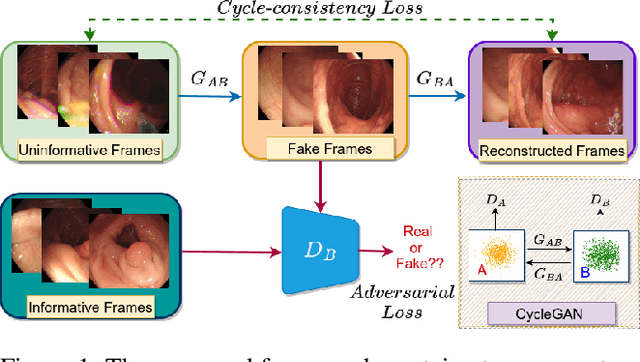
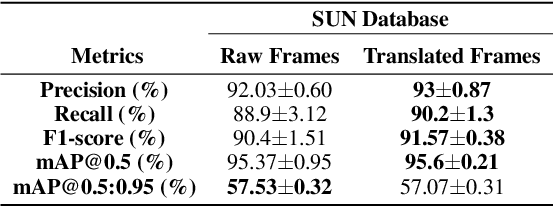
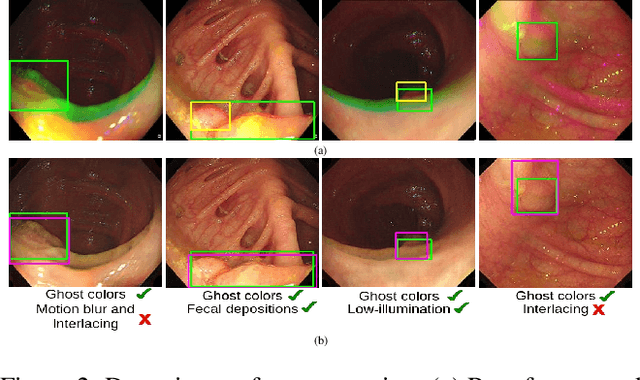
Abstract:Various artifacts, such as ghost colors, interlacing, and motion blur, hinder diagnosing colorectal cancer (CRC) from videos acquired during colonoscopy. The frames containing these artifacts are called uninformative frames and are present in large proportions in colonoscopy videos. To alleviate the impact of artifacts, we propose an adversarial network based framework to convert uninformative frames to clinically relevant frames. We examine the effectiveness of the proposed approach by evaluating the translated frames for polyp detection using YOLOv5. Preliminary results present improved detection performance along with elegant qualitative outcomes. We also examine the failure cases to determine the directions for future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge