Neethi Dasu
L2GNet: Optimal Local-to-Global Representation of Anatomical Structures for Generalized Medical Image Segmentation
Feb 06, 2025



Abstract:Continuous Latent Space (CLS) and Discrete Latent Space (DLS) models, like AttnUNet and VQUNet, have excelled in medical image segmentation. In contrast, Synergistic Continuous and Discrete Latent Space (CDLS) models show promise in handling fine and coarse-grained information. However, they struggle with modeling long-range dependencies. CLS or CDLS-based models, such as TransUNet or SynergyNet are adept at capturing long-range dependencies. Since they rely heavily on feature pooling or aggregation using self-attention, they may capture dependencies among redundant regions. This hinders comprehension of anatomical structure content, poses challenges in modeling intra-class and inter-class dependencies, increases false negatives and compromises generalization. Addressing these issues, we propose L2GNet, which learns global dependencies by relating discrete codes obtained from DLS using optimal transport and aligning codes on a trainable reference. L2GNet achieves discriminative on-the-fly representation learning without an additional weight matrix in self-attention models, making it computationally efficient for medical applications. Extensive experiments on multi-organ segmentation and cardiac datasets demonstrate L2GNet's superiority over state-of-the-art methods, including the CDLS method SynergyNet, offering an novel approach to enhance deep learning models' performance in medical image analysis.
GastroVision: A Multi-class Endoscopy Image Dataset for Computer Aided Gastrointestinal Disease Detection
Jul 16, 2023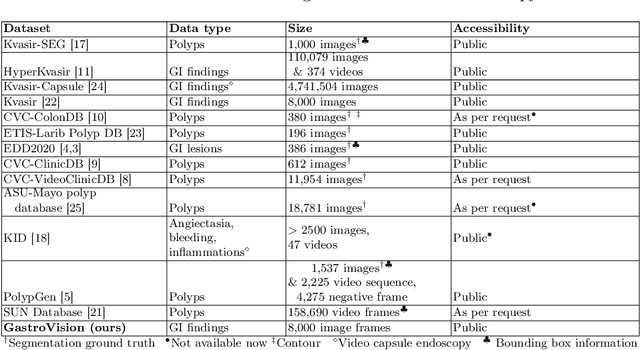
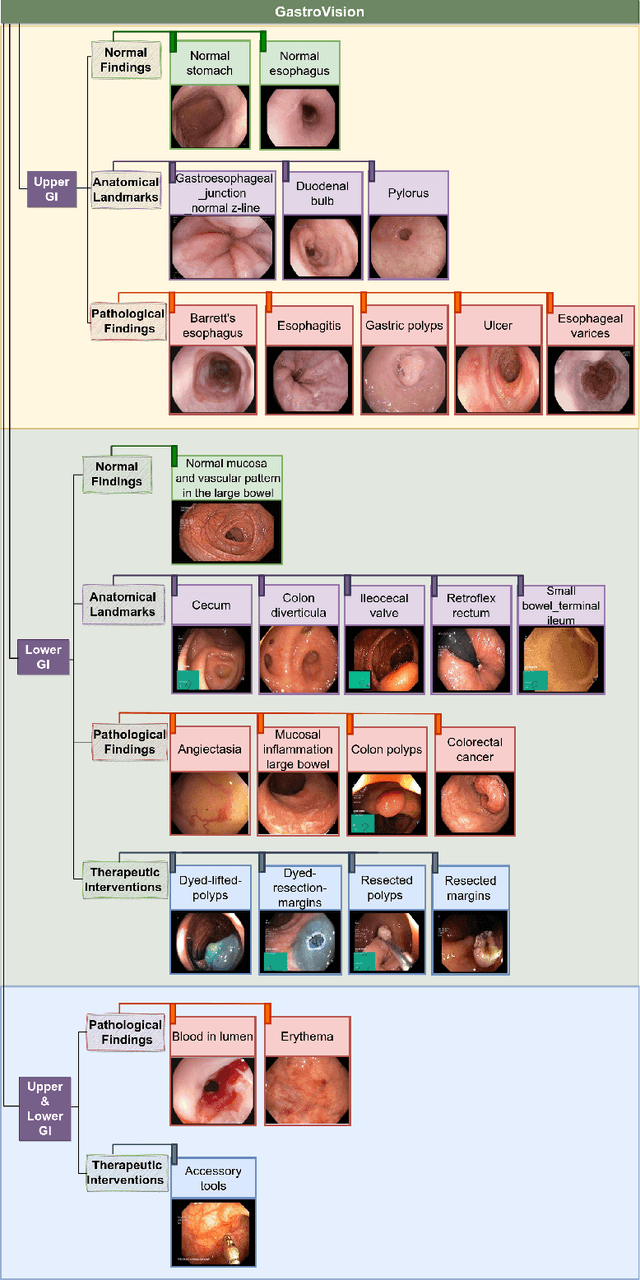
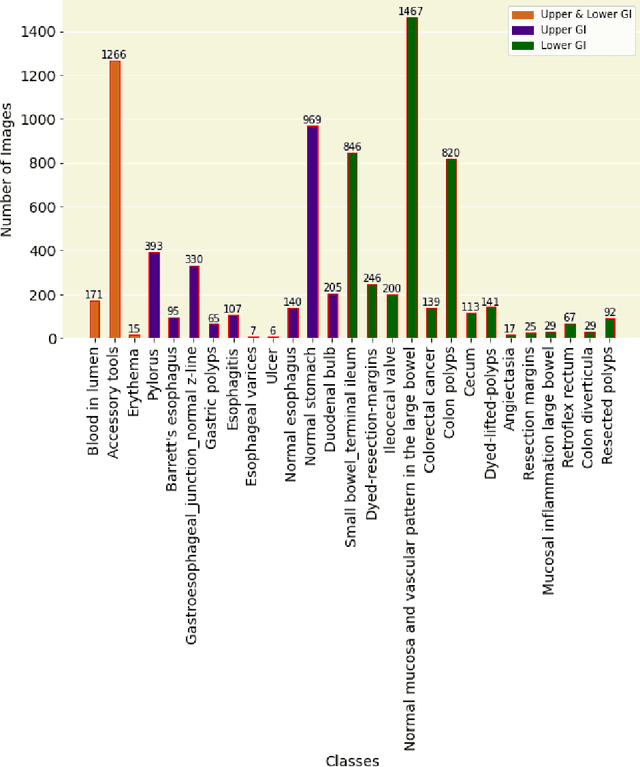
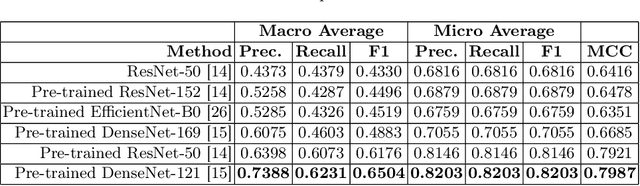
Abstract:Integrating real-time artificial intelligence (AI) systems in clinical practices faces challenges such as scalability and acceptance. These challenges include data availability, biased outcomes, data quality, lack of transparency, and underperformance on unseen datasets from different distributions. The scarcity of large-scale, precisely labeled, and diverse datasets are the major challenge for clinical integration. This scarcity is also due to the legal restrictions and extensive manual efforts required for accurate annotations from clinicians. To address these challenges, we present GastroVision, a multi-center open-access gastrointestinal (GI) endoscopy dataset that includes different anatomical landmarks, pathological abnormalities, polyp removal cases and normal findings (a total of 24 classes) from the GI tract. The dataset comprises 8,000 images acquired from B{\ae}rum Hospital in Norway and Karolinska University in Sweden and was annotated and verified by experienced GI endoscopists. Furthermore, we validate the significance of our dataset with extensive benchmarking based on the popular deep learning based baseline models. We believe our dataset can facilitate the development of AI-based algorithms for GI disease detection and classification. Our dataset is available at https://osf.io/84e7f/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge