Zihan Wan
Detecting Schizophrenia with 3D Structural Brain MRI Using Deep Learning
Jul 07, 2022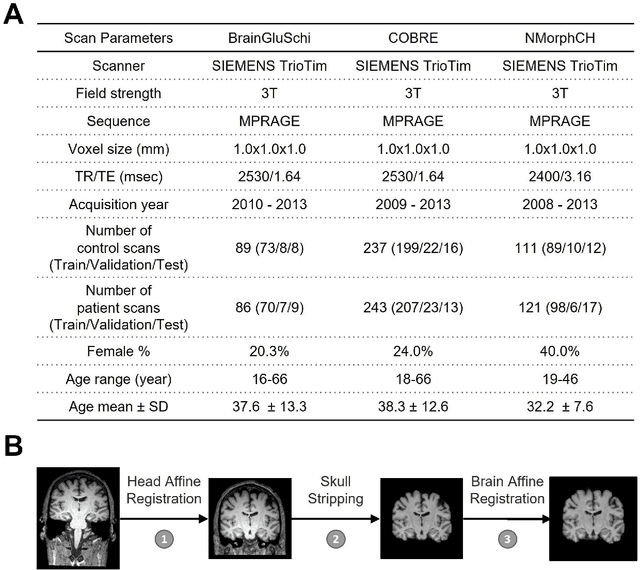
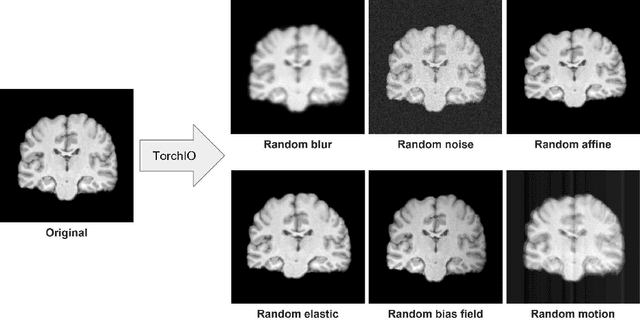
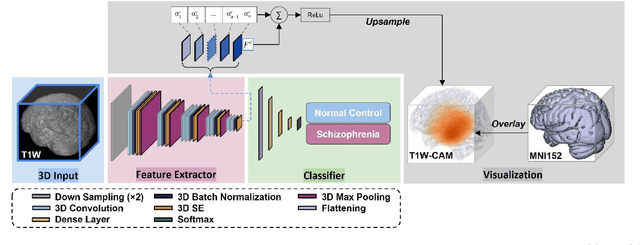
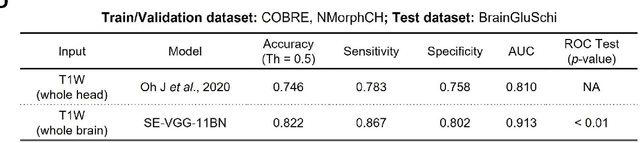
Abstract:Schizophrenia is a chronic neuropsychiatric disorder that causes distinct structural alterations within the brain. We hypothesize that deep learning applied to a structural neuroimaging dataset could detect disease-related alteration and improve classification and diagnostic accuracy. We tested this hypothesis using a single, widely available, and conventional T1-weighted MRI scan, from which we extracted the 3D whole-brain structure using standard post-processing methods. A deep learning model was then developed, optimized, and evaluated on three open datasets with T1-weighted MRI scans of patients with schizophrenia. Our proposed model outperformed the benchmark model, which was also trained with structural MR images using a 3D CNN architecture. Our model is capable of almost perfectly (area under the ROC curve = 0.987) distinguishing schizophrenia patients from healthy controls on unseen structural MRI scans. Regional analysis localized subcortical regions and ventricles as the most predictive brain regions. Subcortical structures serve a pivotal role in cognitive, affective, and social functions in humans, and structural abnormalities of these regions have been associated with schizophrenia. Our finding corroborates that schizophrenia is associated with widespread alterations in subcortical brain structure and the subcortical structural information provides prominent features in diagnostic classification. Together, these results further demonstrate the potential of deep learning to improve schizophrenia diagnosis and identify its structural neuroimaging signatures from a single, standard T1-weighted brain MRI.
Improving Across-Dataset Brain Tissue Segmentation Using Transformer
Jan 21, 2022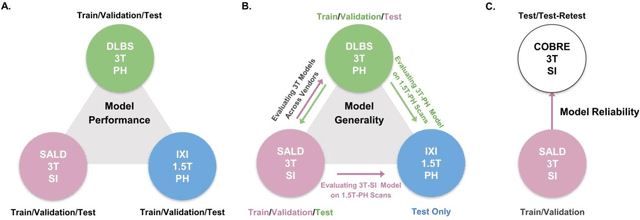
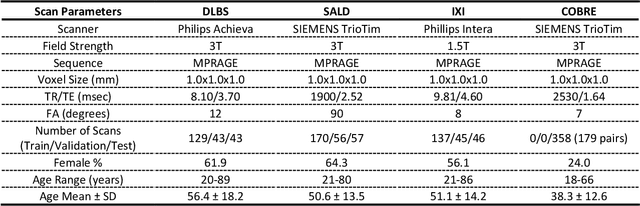
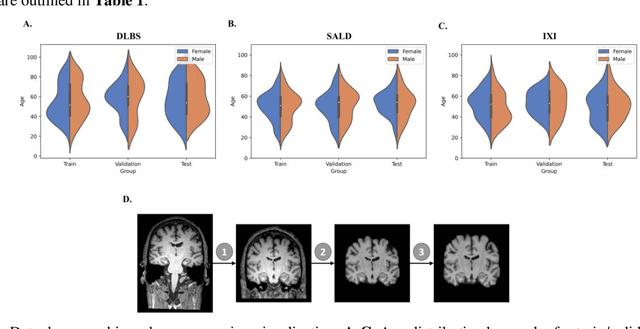
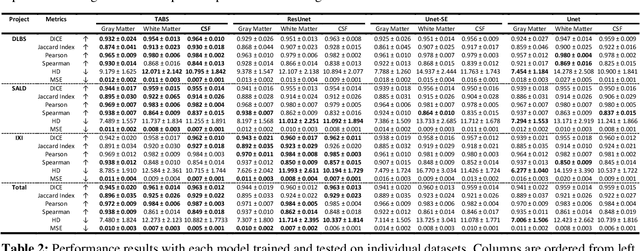
Abstract:Brain tissue segmentation has demonstrated great utility in quantifying MRI data through Voxel-Based Morphometry and highlighting subtle structural changes associated with various conditions within the brain. However, manual segmentation is highly labor-intensive, and automated approaches have struggled due to properties inherent to MRI acquisition, leaving a great need for an effective segmentation tool. Despite the recent success of deep convolutional neural networks (CNNs) for brain tissue segmentation, many such solutions do not generalize well to new datasets, which is critical for a reliable solution. Transformers have demonstrated success in natural image segmentation and have recently been applied to 3D medical image segmentation tasks due to their ability to capture long-distance relationships in the input where the local receptive fields of CNNs struggle. This study introduces a novel CNN-Transformer hybrid architecture designed for brain tissue segmentation. We validate our model's performance across four multi-site T1w MRI datasets, covering different vendors, field strengths, scan parameters, time points, and neuropsychiatric conditions. In all situations, our model achieved the greatest generality and reliability. Out method is inherently robust and can serve as a valuable tool for brain-related T1w MRI studies. The code for the TABS network is available at: https://github.com/raovish6/TABS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge