Jean-Christophe Olivo-Marin
AIQ
SINETRA: a Versatile Framework for Evaluating Single Neuron Tracking in Behaving Animals
Nov 14, 2024Abstract:Accurately tracking neuronal activity in behaving animals presents significant challenges due to complex motions and background noise. The lack of annotated datasets limits the evaluation and improvement of such tracking algorithms. To address this, we developed SINETRA, a versatile simulator that generates synthetic tracking data for particles on a deformable background, closely mimicking live animal recordings. This simulator produces annotated 2D and 3D videos that reflect the intricate movements seen in behaving animals like Hydra Vulgaris. We evaluated four state-of-the-art tracking algorithms highlighting the current limitations of these methods in challenging scenarios and paving the way for improved cell tracking techniques in dynamic biological systems.
Deep ContourFlow: Advancing Active Contours with Deep Learning
Jul 15, 2024

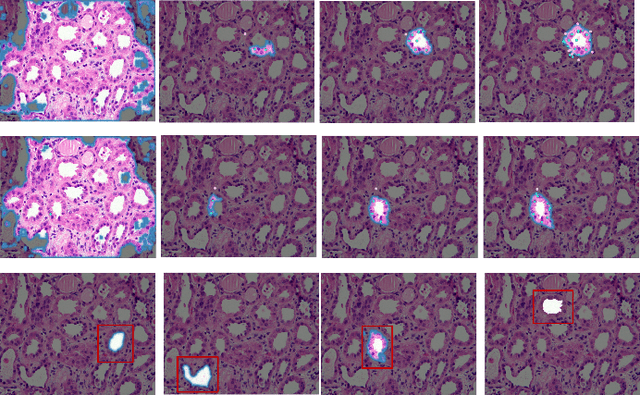
Abstract:This paper introduces a novel approach that combines unsupervised active contour models with deep learning for robust and adaptive image segmentation. Indeed, traditional active contours, provide a flexible framework for contour evolution and learning offers the capacity to learn intricate features and patterns directly from raw data. Our proposed methodology leverages the strengths of both paradigms, presenting a framework for both unsupervised and one-shot approaches for image segmentation. It is capable of capturing complex object boundaries without the need for extensive labeled training data. This is particularly required in histology, a field facing a significant shortage of annotations due to the challenging and time-consuming nature of the annotation process. We illustrate and compare our results to state of the art methods on a histology dataset and show significant improvements.
Scribble-based fast weak-supervision and interactive corrections for segmenting whole slide images
Feb 13, 2024



Abstract:This paper proposes a dynamic interactive and weakly supervised segmentation method with minimal user interactions to address two major challenges in the segmentation of whole slide histopathology images. First, the lack of hand-annotated datasets to train algorithms. Second, the lack of interactive paradigms to enable a dialogue between the pathologist and the machine, which can be a major obstacle for use in clinical routine. We therefore propose a fast and user oriented method to bridge this gap by giving the pathologist control over the final result while limiting the number of interactions needed to achieve a good result (over 90\% on all our metrics with only 4 correction scribbles).
JDLL: A library to run Deep Learning models on Java bioimage informatics platforms
Jun 07, 2023Abstract:We present JDLL, an agile Java library that offers a comprehensive toolset/API to unify the development of high-end applications of DL for bioimage analysis and to streamline their installation and maintenance. JDLL provides all the functions required to consume DL models seamlessly, without being burdened by the configuration of the Python-based DL frameworks, within Java bioimage informatics platforms. Moreover, it allows the deployment of pre-trained models in the Bioimage Model Zoo (BMZ) by shipping the logic to connect to the BMZ website, download and run a selected model inference.
From Nano to Macro: Overview of the IEEE Bio Image and Signal Processing Technical Committee
Oct 31, 2022

Abstract:The Bio Image and Signal Processing (BISP) Technical Committee (TC) of the IEEE Signal Processing Society (SPS) promotes activities within the broad technical field of biomedical image and signal processing. Areas of interest include medical and biological imaging, digital pathology, molecular imaging, microscopy, and associated computational imaging, image analysis, and image-guided treatment, alongside physiological signal processing, computational biology, and bioinformatics. BISP has 40 members and covers a wide range of EDICS, including CIS-MI: Medical Imaging, BIO-MIA: Medical Image Analysis, BIO-BI: Biological Imaging, BIO: Biomedical Signal Processing, BIO-BCI: Brain/Human-Computer Interfaces, and BIO-INFR: Bioinformatics. BISP plays a central role in the organization of the IEEE International Symposium on Biomedical Imaging (ISBI) and contributes to the technical sessions at the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), and the IEEE International Conference on Image Processing (ICIP). In this paper, we provide a brief history of the TC, review the technological and methodological contributions its community delivered, and highlight promising new directions we anticipate.
Learning to segment clustered amoeboid cells from brightfield microscopy via multi-task learning with adaptive weight selection
May 19, 2020
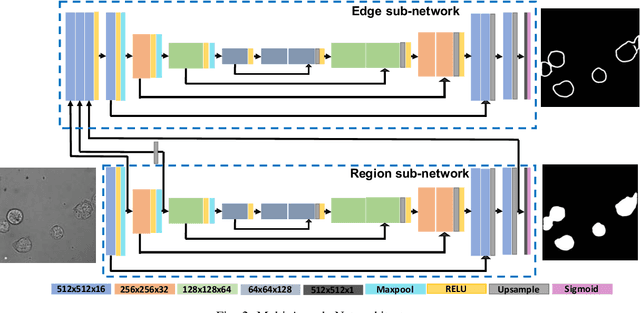


Abstract:Detecting and segmenting individual cells from microscopy images is critical to various life science applications. Traditional cell segmentation tools are often ill-suited for applications in brightfield microscopy due to poor contrast and intensity heterogeneity, and only a small subset are applicable to segment cells in a cluster. In this regard, we introduce a novel supervised technique for cell segmentation in a multi-task learning paradigm. A combination of a multi-task loss, based on the region and cell boundary detection, is employed for an improved prediction efficiency of the network. The learning problem is posed in a novel min-max framework which enables adaptive estimation of the hyper-parameters in an automatic fashion. The region and cell boundary predictions are combined via morphological operations and active contour model to segment individual cells. The proposed methodology is particularly suited to segment touching cells from brightfield microscopy images without manual interventions. Quantitatively, we observe an overall Dice score of 0.93 on the validation set, which is an improvement of over 15.9% on a recent unsupervised method, and outperforms the popular supervised U-net algorithm by at least $5.8\%$ on average.
Compressed Sensing with off-axis frequency-shifting holography
Apr 29, 2010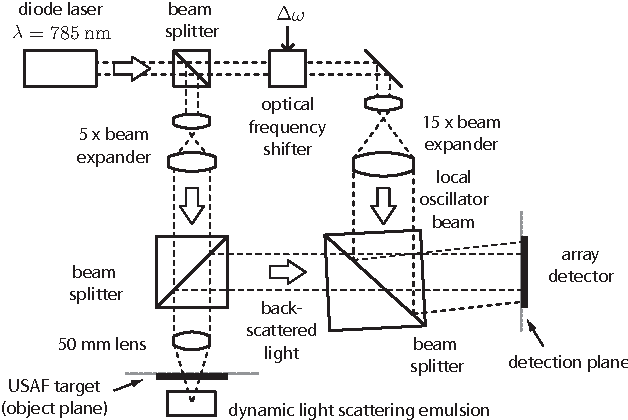
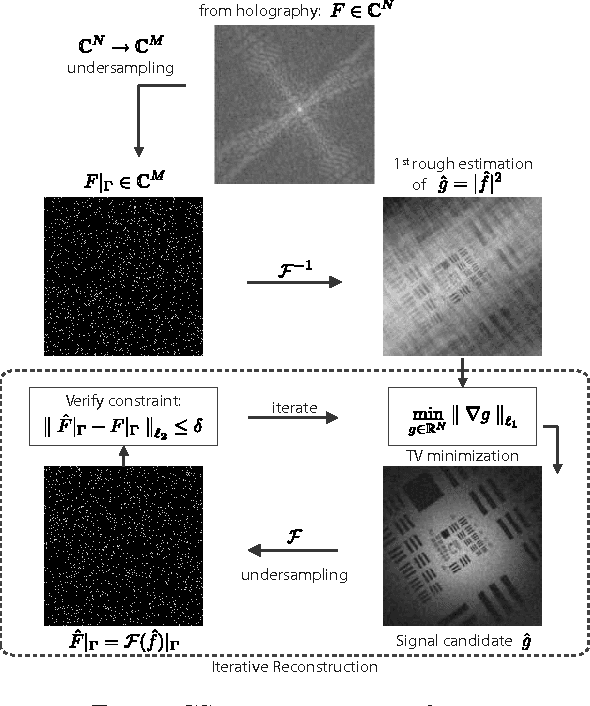
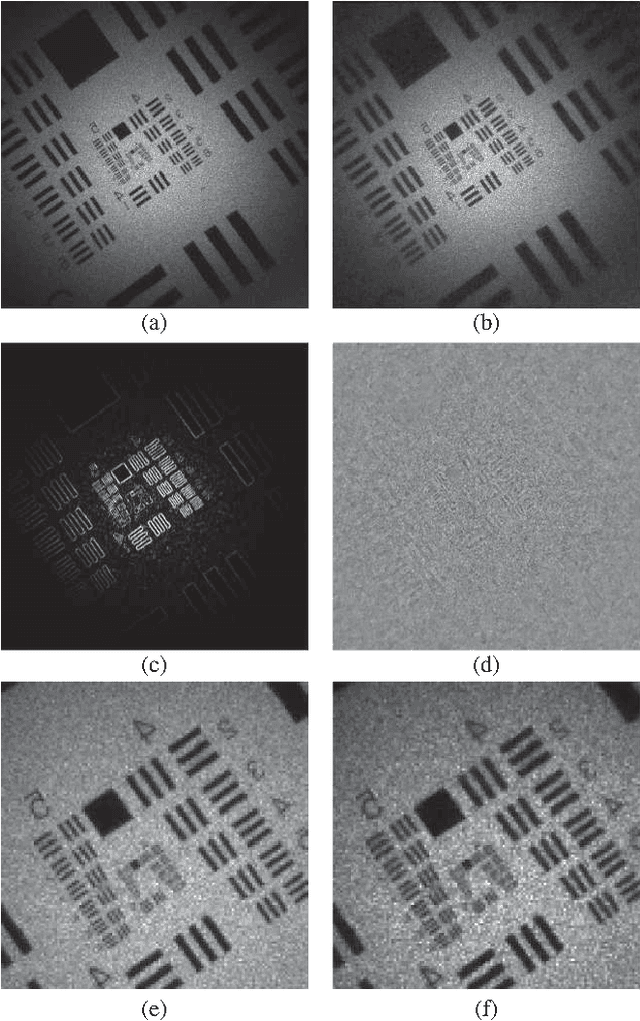
Abstract:This work reveals an experimental microscopy acquisition scheme successfully combining Compressed Sensing (CS) and digital holography in off-axis and frequency-shifting conditions. CS is a recent data acquisition theory involving signal reconstruction from randomly undersampled measurements, exploiting the fact that most images present some compact structure and redundancy. We propose a genuine CS-based imaging scheme for sparse gradient images, acquiring a diffraction map of the optical field with holographic microscopy and recovering the signal from as little as 7% of random measurements. We report experimental results demonstrating how CS can lead to an elegant and effective way to reconstruct images, opening the door for new microscopy applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge