Arrate Muñoz-Barrutia
Bioengineering Department, Universidad Carlos III de Madrid, Instituto de Investigación Sanitaria Gregorio Marañón
OREHAS: A fully automated deep-learning pipeline for volumetric endolymphatic hydrops quantification in MRI
Jan 26, 2026Abstract:We present OREHAS (Optimized Recognition & Evaluation of volumetric Hydrops in the Auditory System), the first fully automatic pipeline for volumetric quantification of endolymphatic hydrops (EH) from routine 3D-SPACE-MRC and 3D-REAL-IR MRI. The system integrates three components -- slice classification, inner ear localization, and sequence-specific segmentation -- into a single workflow that computes per-ear endolymphatic-to-vestibular volume ratios (ELR) directly from whole MRI volumes, eliminating the need for manual intervention. Trained with only 3 to 6 annotated slices per patient, OREHAS generalized effectively to full 3D volumes, achieving Dice scores of 0.90 for SPACE-MRC and 0.75 for REAL-IR. In an external validation cohort with complete manual annotations, OREHAS closely matched expert ground truth (VSI = 74.3%) and substantially outperformed the clinical syngo.via software (VSI = 42.5%), which tended to overestimate endolymphatic volumes. Across 19 test patients, vestibular measurements from OREHAS were consistent with syngo.via, while endolymphatic volumes were systematically smaller and more physiologically realistic. These results show that reliable and reproducible EH quantification can be achieved from standard MRI using limited supervision. By combining efficient deep-learning-based segmentation with a clinically aligned volumetric workflow, OREHAS reduces operator dependence, ensures methodological consistency. Besides, the results are compatible with established imaging protocols. The approach provides a robust foundation for large-scale studies and for recalibrating clinical diagnostic thresholds based on accurate volumetric measurements of the inner ear.
Multimodal Posterior Sampling-based Uncertainty in PD-L1 Segmentation from H&E Images
Nov 14, 2025



Abstract:Accurate assessment of PD-L1 expression is critical for guiding immunotherapy, yet current immunohistochemistry (IHC) based methods are resource-intensive. We present nnUNet-B: a Bayesian segmentation framework that infers PD-L1 expression directly from H&E-stained histology images using Multimodal Posterior Sampling (MPS). Built upon nnUNet-v2, our method samples diverse model checkpoints during cyclic training to approximate the posterior, enabling both accurate segmentation and epistemic uncertainty estimation via entropy and standard deviation. Evaluated on a dataset of lung squamous cell carcinoma, our approach achieves competitive performance against established baselines with mean Dice Score and mean IoU of 0.805 and 0.709, respectively, while providing pixel-wise uncertainty maps. Uncertainty estimates show strong correlation with segmentation error, though calibration remains imperfect. These results suggest that uncertainty-aware H&E-based PD-L1 prediction is a promising step toward scalable, interpretable biomarker assessment in clinical workflows.
Alzheimer's disease detection in PSG signals
Apr 04, 2024Abstract:Alzheimer's disease (AD) and sleep disorders exhibit a close association, where disruptions in sleep patterns often precede the onset of Mild Cognitive Impairment (MCI) and early-stage AD. This study delves into the potential of utilizing sleep-related electroencephalography (EEG) signals acquired through polysomnography (PSG) for the early detection of AD. Our primary focus is on exploring semi-supervised Deep Learning techniques for the classification of EEG signals due to the clinical scenario characterized by the limited data availability. The methodology entails testing and comparing the performance of semi-supervised SMATE and TapNet models, benchmarked against the supervised XCM model, and unsupervised Hidden Markov Models (HMMs). The study highlights the significance of spatial and temporal analysis capabilities, conducting independent analyses of each sleep stage. Results demonstrate the effectiveness of SMATE in leveraging limited labeled data, achieving stable metrics across all sleep stages, and reaching 90% accuracy in its supervised form. Comparative analyses reveal SMATE's superior performance over TapNet and HMM, while XCM excels in supervised scenarios with an accuracy range of 92 - 94%. These findings underscore the potential of semi-supervised models in early AD detection, particularly in overcoming the challenges associated with the scarcity of labeled data. Ablation tests affirm the critical role of spatio-temporal feature extraction in semi-supervised predictive performance, and t-SNE visualizations validate the model's proficiency in distinguishing AD patterns. Overall, this research contributes to the advancement of AD detection through innovative Deep Learning approaches, highlighting the crucial role of semi-supervised learning in addressing data limitations.
MIFA: Metadata, Incentives, Formats, and Accessibility guidelines to improve the reuse of AI datasets for bioimage analysis
Nov 22, 2023



Abstract:Artificial Intelligence methods are powerful tools for biological image analysis and processing. High-quality annotated images are key to training and developing new methods, but access to such data is often hindered by the lack of standards for sharing datasets. We brought together community experts in a workshop to develop guidelines to improve the reuse of bioimages and annotations for AI applications. These include standards on data formats, metadata, data presentation and sharing, and incentives to generate new datasets. We are positive that the MIFA (Metadata, Incentives, Formats, and Accessibility) recommendations will accelerate the development of AI tools for bioimage analysis by facilitating access to high quality training data.
BioImage.IO Chatbot: A Personalized Assistant for BioImage Analysis Augmented by Community Knowledge Base
Oct 31, 2023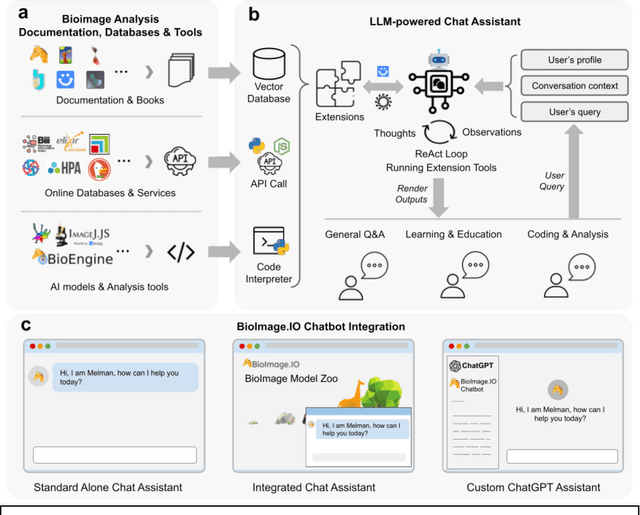
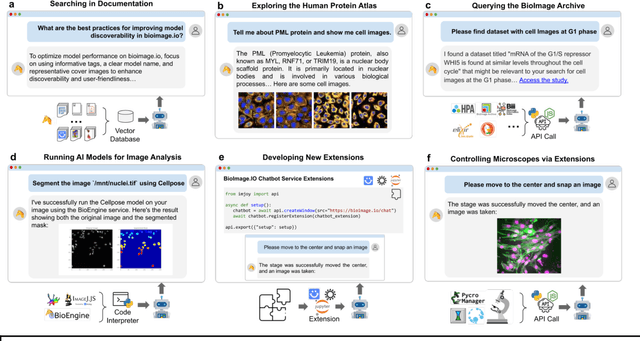
Abstract:The rapidly expanding landscape of bioimage analysis tools presents a navigational challenge for both experts and newcomers. Traditional search methods often fall short in assisting users in this complex environment. To address this, we introduce the BioImage$.$IO Chatbot, an AI-driven conversational assistant tailored for the bioimage community. Built upon large language models, this chatbot provides personalized, context-aware answers by aggregating and interpreting information from diverse databases, tool-specific documentation, and structured data sources. Enhanced by a community-contributed knowledge base and fine-tuned retrieval methods, the BioImage$.$IO Chatbot offers not just a personalized interaction but also a knowledge-enriched, context-aware experience. It fundamentally transforms the way biologists, bioimage analysts, and developers navigate and utilize advanced bioimage analysis tools, setting a new standard for community-driven, accessible scientific research.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023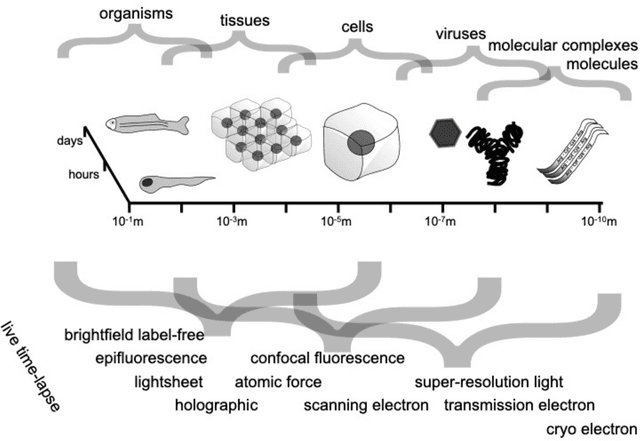
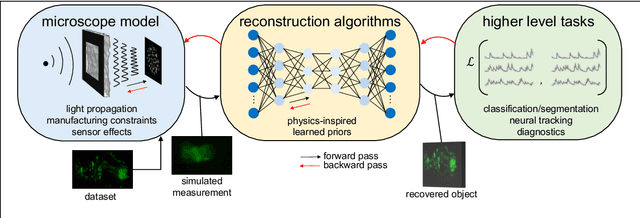
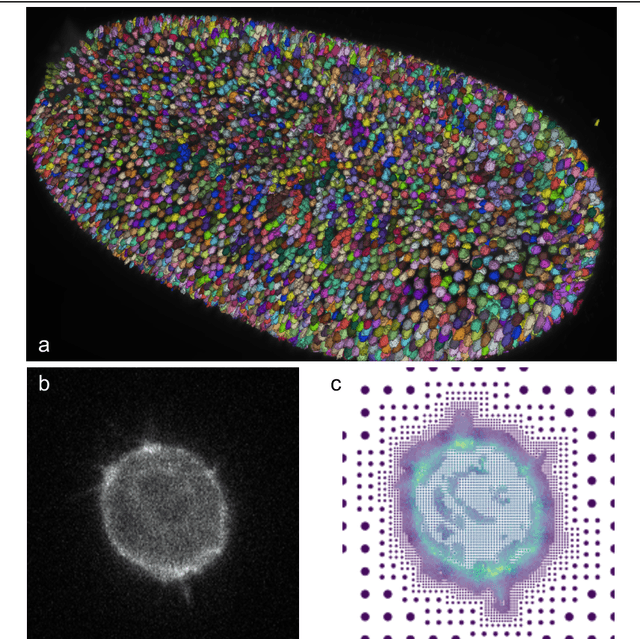
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
ABANICCO: A New Color Space for Multi-Label Pixel Classification and Color Segmentation
Nov 15, 2022Abstract:In any computer vision task involving color images, a necessary step is classifying pixels according to color and segmenting the respective areas. However, the development of methods able to successfully complete this task has proven challenging, mainly due to the gap between human color perception, linguistic color terms, and digital representation. In this paper, we propose a novel method combining geometric analysis of color theory, fuzzy color spaces, and multi-label systems for the automatic classification of pixels according to 12 standard color categories (Green, Yellow, Light Orange, Deep Orange, Red, Pink, Purple, Ultramarine, Blue, Teal, Brown, and Neutral). Moreover, we present a robust, unsupervised, unbiased strategy for color naming based on statistics and color theory. ABANICCO was tested against the state of the art in color classification and with the standarized ISCC-NBS color system, providing accurate classification and a standard, easily understandable alternative for hue naming recognizable by humans and machines. We expect this solution to become the base to successfully tackle a myriad of problems in all fields of computer vision, such as region characterization, histopathology analysis, fire detection, product quality prediction, object description, and hyperspectral imaging.
From Nano to Macro: Overview of the IEEE Bio Image and Signal Processing Technical Committee
Oct 31, 2022

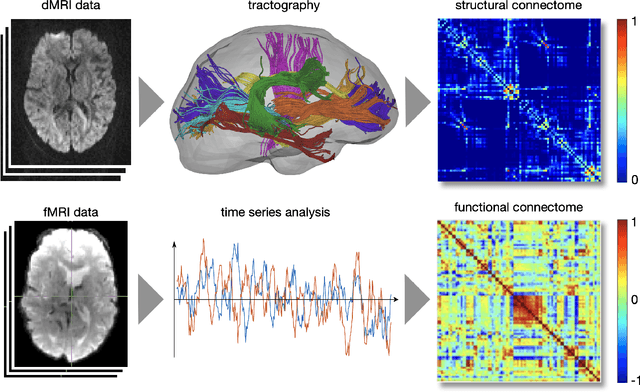
Abstract:The Bio Image and Signal Processing (BISP) Technical Committee (TC) of the IEEE Signal Processing Society (SPS) promotes activities within the broad technical field of biomedical image and signal processing. Areas of interest include medical and biological imaging, digital pathology, molecular imaging, microscopy, and associated computational imaging, image analysis, and image-guided treatment, alongside physiological signal processing, computational biology, and bioinformatics. BISP has 40 members and covers a wide range of EDICS, including CIS-MI: Medical Imaging, BIO-MIA: Medical Image Analysis, BIO-BI: Biological Imaging, BIO: Biomedical Signal Processing, BIO-BCI: Brain/Human-Computer Interfaces, and BIO-INFR: Bioinformatics. BISP plays a central role in the organization of the IEEE International Symposium on Biomedical Imaging (ISBI) and contributes to the technical sessions at the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), and the IEEE International Conference on Image Processing (ICIP). In this paper, we provide a brief history of the TC, review the technological and methodological contributions its community delivered, and highlight promising new directions we anticipate.
Translational Lung Imaging Analysis Through Disentangled Representations
Mar 03, 2022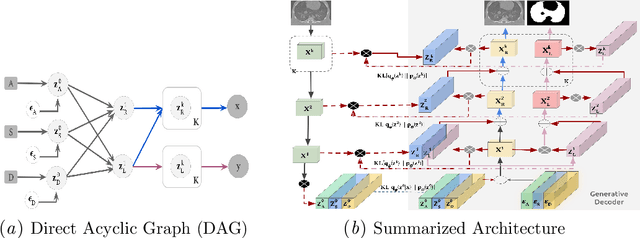
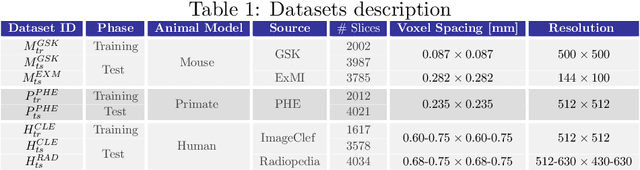
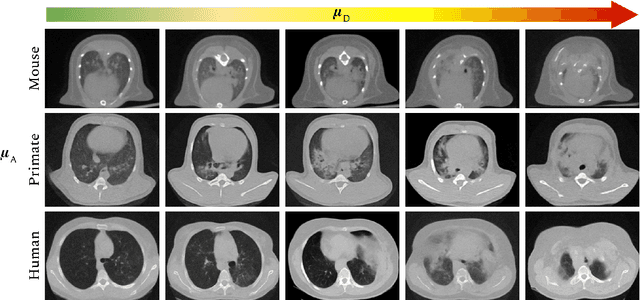

Abstract:The development of new treatments often requires clinical trials with translational animal models using (pre)-clinical imaging to characterize inter-species pathological processes. Deep Learning (DL) models are commonly used to automate retrieving relevant information from the images. Nevertheless, they typically suffer from low generability and explainability as a product of their entangled design, resulting in a specific DL model per animal model. Consequently, it is not possible to take advantage of the high capacity of DL to discover statistical relationships from inter-species images. To alleviate this problem, in this work, we present a model capable of extracting disentangled information from images of different animal models and the mechanisms that generate the images. Our method is located at the intersection between deep generative models, disentanglement and causal representation learning. It is optimized from images of pathological lung infected by Tuberculosis and is able: a) from an input slice, infer its position in a volume, the animal model to which it belongs, the damage present and even more, generate a mask covering the whole lung (similar overlap measures to the nnU-Net), b) generate realistic lung images by setting the above variables and c) generate counterfactual images, namely, healthy versions of a damaged input slice.
Deep learning based domain adaptation for mitochondria segmentation on EM volumes
Feb 22, 2022



Abstract:Accurate segmentation of electron microscopy (EM) volumes of the brain is essential to characterize neuronal structures at a cell or organelle level. While supervised deep learning methods have led to major breakthroughs in that direction during the past years, they usually require large amounts of annotated data to be trained, and perform poorly on other data acquired under similar experimental and imaging conditions. This is a problem known as domain adaptation, since models that learned from a sample distribution (or source domain) struggle to maintain their performance on samples extracted from a different distribution or target domain. In this work, we address the complex case of deep learning based domain adaptation for mitochondria segmentation across EM datasets from different tissues and species. We present three unsupervised domain adaptation strategies to improve mitochondria segmentation in the target domain based on (1) state-of-the-art style transfer between images of both domains; (2) self-supervised learning to pre-train a model using unlabeled source and target images, and then fine-tune it only with the source labels; and (3) multi-task neural network architectures trained end-to-end with both labeled and unlabeled images. Additionally, we propose a new training stopping criterion based on morphological priors obtained exclusively in the source domain. We carried out all possible cross-dataset experiments using three publicly available EM datasets. We evaluated our proposed strategies on the mitochondria semantic labels predicted on the target datasets. The methods introduced here outperform the baseline methods and compare favorably to the state of the art. In the absence of validation labels, monitoring our proposed morphology-based metric is an intuitive and effective way to stop the training process and select in average optimal models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge