Caterina Fuster-Barceló
Are Vision Foundation Models Foundational for Electron Microscopy Image Segmentation?
Feb 09, 2026Abstract:Although vision foundation models (VFMs) are increasingly reused for biomedical image analysis, it remains unclear whether the latent representations they provide are general enough to support effective transfer and reuse across heterogeneous microscopy image datasets. Here, we study this question for the problem of mitochondria segmentation in electron microscopy (EM) images, using two popular public EM datasets (Lucchi++ and VNC) and three recent representative VFMs (DINOv2, DINOv3, and OpenCLIP). We evaluate two practical model adaptation regimes: a frozen-backbone setting in which only a lightweight segmentation head is trained on top of the VFM, and parameter-efficient fine-tuning (PEFT) via Low-Rank Adaptation (LoRA) in which the VFM is fine-tuned in a targeted manner to a specific dataset. Across all backbones, we observe that training on a single EM dataset yields good segmentation performance (quantified as foreground Intersection-over-Union), and that LoRA consistently improves in-domain performance. In contrast, training on multiple EM datasets leads to severe performance degradation for all models considered, with only marginal gains from PEFT. Exploration of the latent representation space through various techniques (PCA, Fréchet Dinov2 distance, and linear probes) reveals a pronounced and persistent domain mismatch between the two considered EM datasets in spite of their visual similarity, which is consistent with the observed failure of paired training. These results suggest that, while VFMs can deliver competitive results for EM segmentation within a single domain under lightweight adaptation, current PEFT strategies are insufficient to obtain a single robust model across heterogeneous EM datasets without additional domain-alignment mechanisms.
OREHAS: A fully automated deep-learning pipeline for volumetric endolymphatic hydrops quantification in MRI
Jan 26, 2026Abstract:We present OREHAS (Optimized Recognition & Evaluation of volumetric Hydrops in the Auditory System), the first fully automatic pipeline for volumetric quantification of endolymphatic hydrops (EH) from routine 3D-SPACE-MRC and 3D-REAL-IR MRI. The system integrates three components -- slice classification, inner ear localization, and sequence-specific segmentation -- into a single workflow that computes per-ear endolymphatic-to-vestibular volume ratios (ELR) directly from whole MRI volumes, eliminating the need for manual intervention. Trained with only 3 to 6 annotated slices per patient, OREHAS generalized effectively to full 3D volumes, achieving Dice scores of 0.90 for SPACE-MRC and 0.75 for REAL-IR. In an external validation cohort with complete manual annotations, OREHAS closely matched expert ground truth (VSI = 74.3%) and substantially outperformed the clinical syngo.via software (VSI = 42.5%), which tended to overestimate endolymphatic volumes. Across 19 test patients, vestibular measurements from OREHAS were consistent with syngo.via, while endolymphatic volumes were systematically smaller and more physiologically realistic. These results show that reliable and reproducible EH quantification can be achieved from standard MRI using limited supervision. By combining efficient deep-learning-based segmentation with a clinically aligned volumetric workflow, OREHAS reduces operator dependence, ensures methodological consistency. Besides, the results are compatible with established imaging protocols. The approach provides a robust foundation for large-scale studies and for recalibrating clinical diagnostic thresholds based on accurate volumetric measurements of the inner ear.
BioImage.IO Chatbot: A Personalized Assistant for BioImage Analysis Augmented by Community Knowledge Base
Oct 31, 2023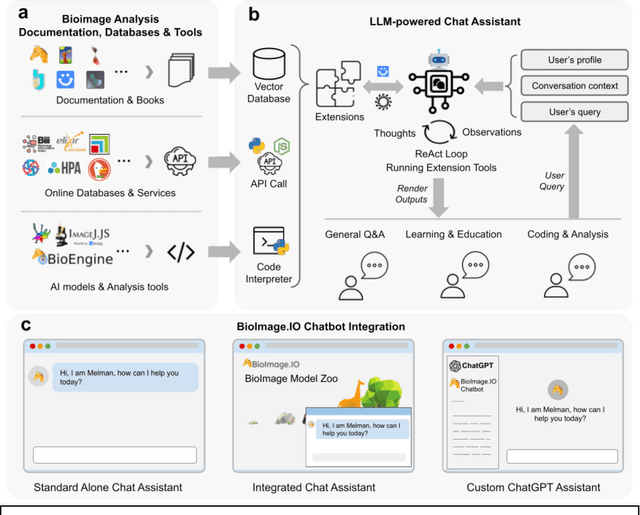
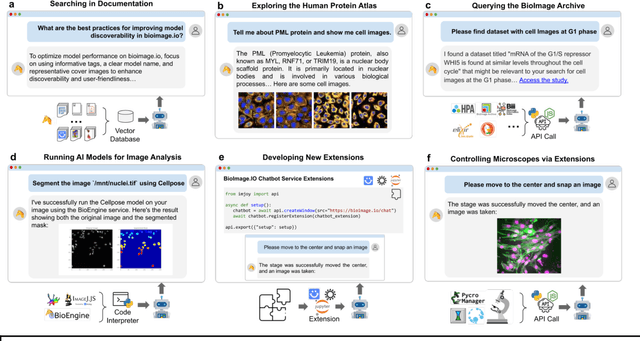
Abstract:The rapidly expanding landscape of bioimage analysis tools presents a navigational challenge for both experts and newcomers. Traditional search methods often fall short in assisting users in this complex environment. To address this, we introduce the BioImage$.$IO Chatbot, an AI-driven conversational assistant tailored for the bioimage community. Built upon large language models, this chatbot provides personalized, context-aware answers by aggregating and interpreting information from diverse databases, tool-specific documentation, and structured data sources. Enhanced by a community-contributed knowledge base and fine-tuned retrieval methods, the BioImage$.$IO Chatbot offers not just a personalized interaction but also a knowledge-enriched, context-aware experience. It fundamentally transforms the way biologists, bioimage analysts, and developers navigate and utilize advanced bioimage analysis tools, setting a new standard for community-driven, accessible scientific research.
Unleashing the Power of Electrocardiograms: A novel approach for Patient Identification in Healthcare Systems with ECG Signals
Feb 13, 2023



Abstract:Over the course of the past two decades, a substantial body of research has substantiated the viability of utilising cardiac signals as a biometric modality. This paper presents a novel approach for patient identification in healthcare systems using electrocardiogram signals. A convolutional neural network is used to classify users based on images extracted from ECG signals. The proposed identification system is evaluated in multiple databases, providing a comprehensive understanding of its potential in real-world scenarios. The impact of Cardiovascular Diseases on generic user identification has been largely overlooked in previous studies. The presented method takes into account the cardiovascular condition of the patients, ensuring that the results obtained are not biased or limited. Furthermore, the results obtained are consistent and reliable, with lower error rates and higher accuracy metrics, as demonstrated through extensive experimentation. All these features make the proposed method a valuable contribution to the field of patient identification in healthcare systems, and make it a strong contender for practical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge