Michael Liebling
From Nano to Macro: Overview of the IEEE Bio Image and Signal Processing Technical Committee
Oct 31, 2022

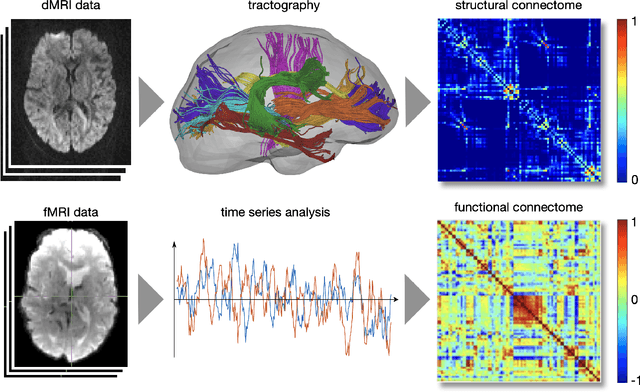
Abstract:The Bio Image and Signal Processing (BISP) Technical Committee (TC) of the IEEE Signal Processing Society (SPS) promotes activities within the broad technical field of biomedical image and signal processing. Areas of interest include medical and biological imaging, digital pathology, molecular imaging, microscopy, and associated computational imaging, image analysis, and image-guided treatment, alongside physiological signal processing, computational biology, and bioinformatics. BISP has 40 members and covers a wide range of EDICS, including CIS-MI: Medical Imaging, BIO-MIA: Medical Image Analysis, BIO-BI: Biological Imaging, BIO: Biomedical Signal Processing, BIO-BCI: Brain/Human-Computer Interfaces, and BIO-INFR: Bioinformatics. BISP plays a central role in the organization of the IEEE International Symposium on Biomedical Imaging (ISBI) and contributes to the technical sessions at the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), and the IEEE International Conference on Image Processing (ICIP). In this paper, we provide a brief history of the TC, review the technological and methodological contributions its community delivered, and highlight promising new directions we anticipate.
Estimating Nonplanar Flow from 2D Motion-blurred Widefield Microscopy Images via Deep Learning
Feb 14, 2021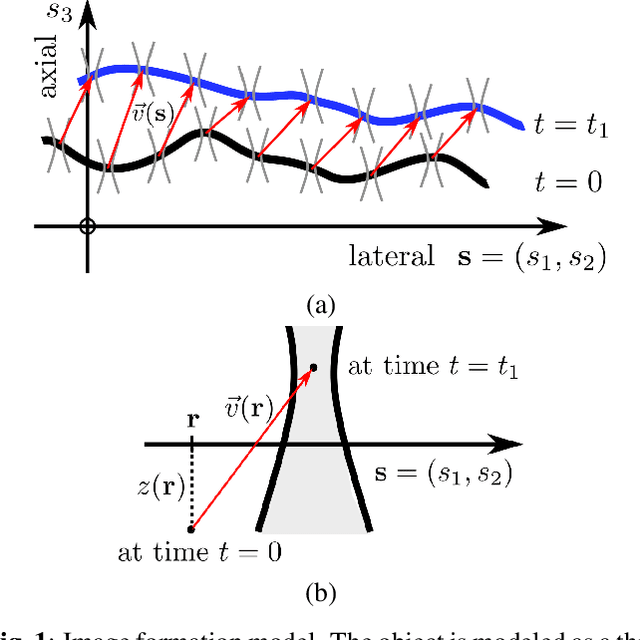
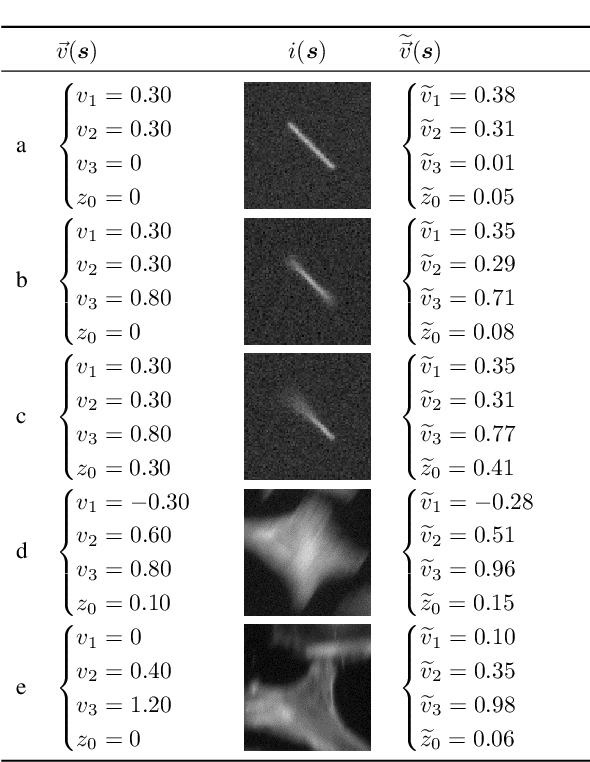

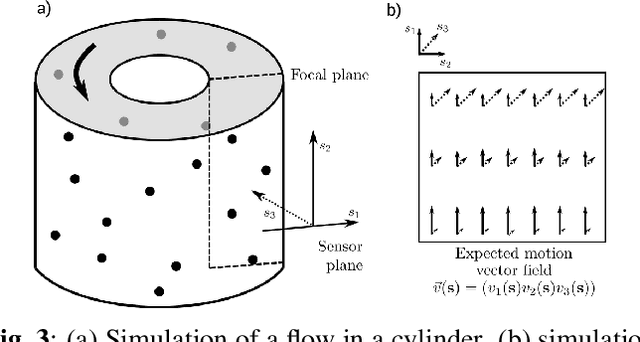
Abstract:Optical flow is a method aimed at predicting the movement velocity of any pixel in the image and is used in medicine and biology to estimate flow of particles in organs or organelles. However, a precise optical flow measurement requires images taken at high speed and low exposure time, which induces phototoxicity due to the increase in illumination power. We are looking here to estimate the three-dimensional movement vector field of moving out-of-plane particles using normal light conditions and a standard microscope camera. We present a method to predict, from a single textured wide-field microscopy image, the movement of out-of-plane particles using the local characteristics of the motion blur. We estimated the velocity vector field from the local estimation of the blur model parameters using an deep neural network and achieved a prediction with a regression coefficient of 0.92 between the ground truth simulated vector field and the output of the network. This method could enable microscopists to gain insights about the dynamic properties of samples without the need for high-speed cameras or high-intensity light exposure.
Spatially-Variant CNN-based Point Spread Function Estimation for Blind Deconvolution and Depth Estimation in Optical Microscopy
Oct 13, 2020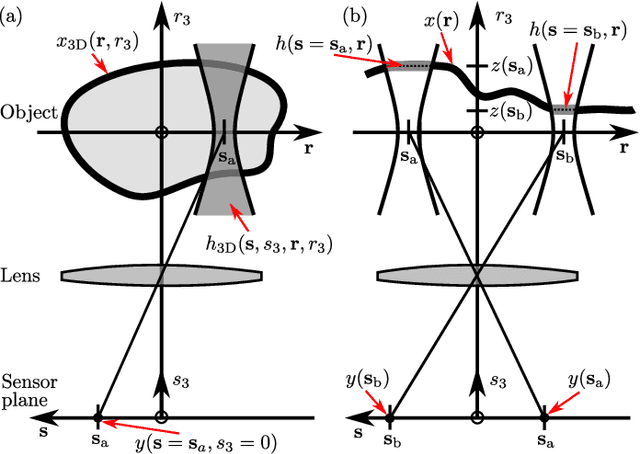
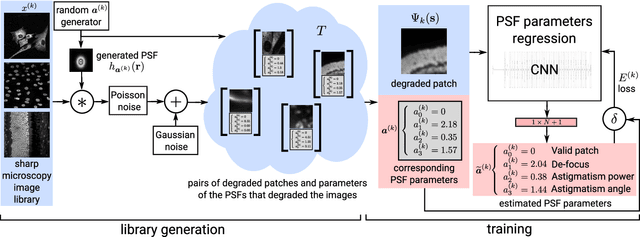
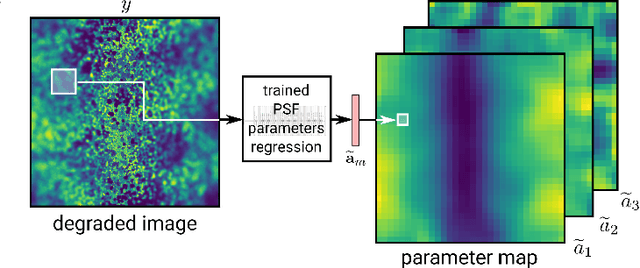
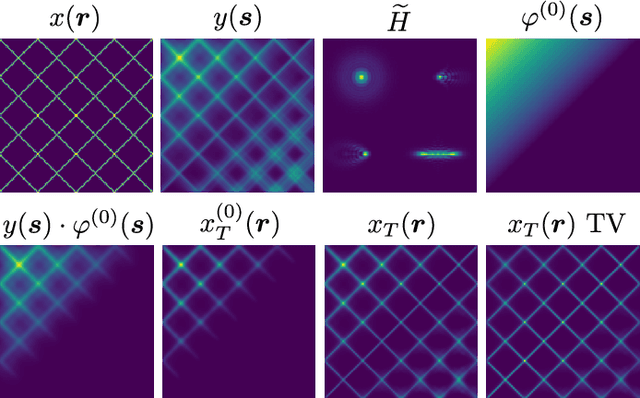
Abstract:Optical microscopy is an essential tool in biology and medicine. Imaging thin, yet non-flat objects in a single shot (without relying on more sophisticated sectioning setups) remains challenging as the shallow depth of field that comes with high-resolution microscopes leads to unsharp image regions and makes depth localization and quantitative image interpretation difficult. Here, we present a method that improves the resolution of light microscopy images of such objects by locally estimating image distortion while jointly estimating object distance to the focal plane. Specifically, we estimate the parameters of a spatially-variant Point-Spread function (PSF) model using a Convolutional Neural Network (CNN), which does not require instrument- or object-specific calibration. Our method recovers PSF parameters from the image itself with up to a squared Pearson correlation coefficient of 0.99 in ideal conditions, while remaining robust to object rotation, illumination variations, or photon noise. When the recovered PSFs are used with a spatially-variant and regularized Richardson-Lucy deconvolution algorithm, we observed up to 2.1 dB better signal-to-noise ratio compared to other blind deconvolution techniques. Following microscope-specific calibration, we further demonstrate that the recovered PSF model parameters permit estimating surface depth with a precision of 2 micrometers and over an extended range when using engineered PSFs. Our method opens up multiple possibilities for enhancing images of non-flat objects with minimal need for a priori knowledge about the optical setup.
* Preprint
Free annotated data for deep learning in microscopy? A hitchhiker's guide
Oct 08, 2020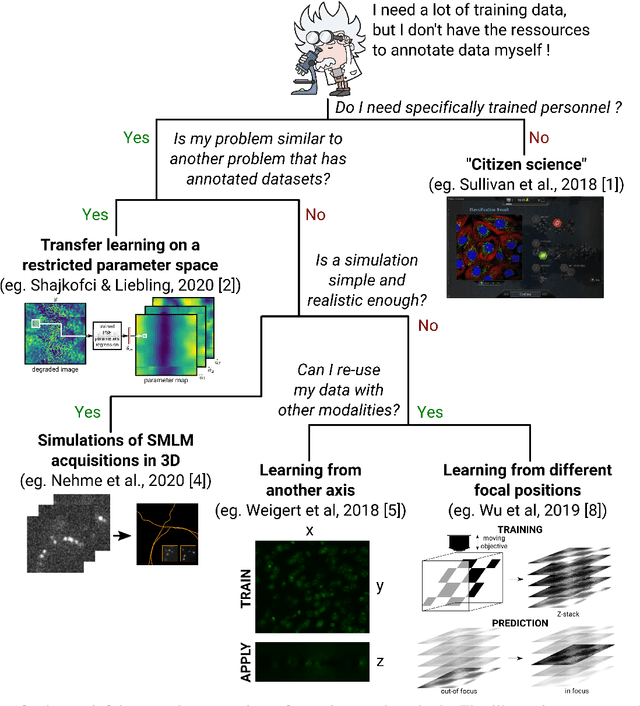
Abstract:In microscopy, the time burden and cost of acquiring and annotating large datasets that many deep learning models take as a prerequisite, often appears to make these methods impractical. Can this requirement for annotated data be relaxed? Is it possible to borrow the knowledge gathered from datasets in other application fields and leverage it for microscopy? Here, we aim to provide an overview of methods that have recently emerged to successfully train learning-based methods in bio-microscopy.
DeepFocus: a Few-Shot Microscope Slide Auto-Focus using a Sample Invariant CNN-based Sharpness Function
Jan 02, 2020

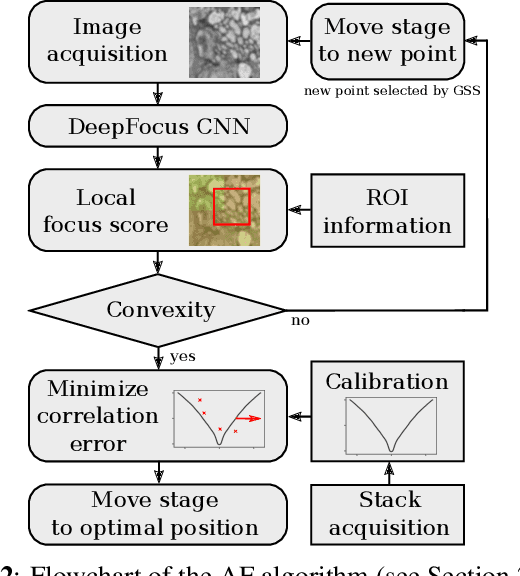
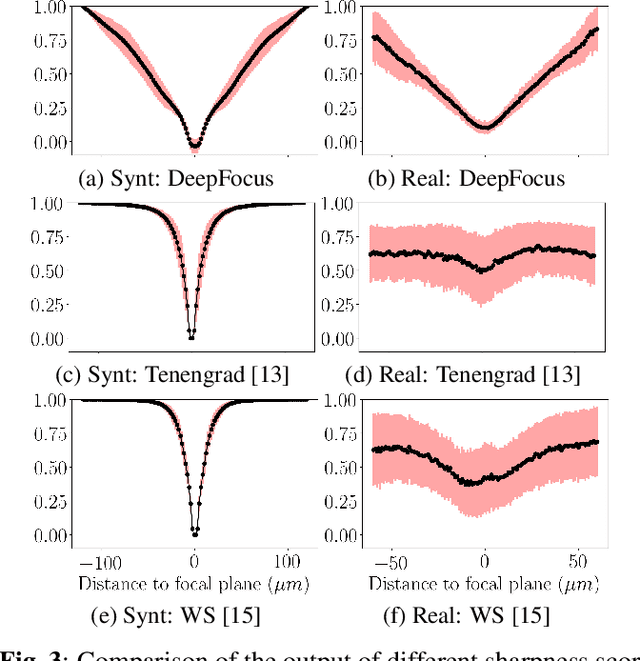
Abstract:Autofocus (AF) methods are extensively used in biomicroscopy, for example to acquire timelapses, where the imaged objects tend to drift out of focus. AD algorithms determine an optimal distance by which to move the sample back into the focal plane. Current hardware-based methods require modifying the microscope and image-based algorithms either rely on many images to converge to the sharpest position or need training data and models specific to each instrument and imaging configuration. Here we propose DeepFocus, an AF method we implemented as a Micro-Manager plugin, and characterize its Convolutional neural network-based sharpness function, which we observed to be depth co-variant and sample-invariant. Sample invariance allows our AF algorithm to converge to an optimal axial position within as few as three iterations using a model trained once for use with a wide range of optical microscopes and a single instrument-dependent calibration stack acquisition of a flat (but arbitrary) textured object. From experiments carried out both on synthetic and experimental data, we observed an average precision, given 3 measured images, of 0.30 +- 0.16 micrometers with a 10x, NA 0.3 objective. We foresee that this performance and low image number will help limit photodamage during acquisitions with light-sensitive samples.
Semi-Blind Spatially-Variant Deconvolution in Optical Microscopy with Local Point Spread Function Estimation By Use Of Convolutional Neural Networks
Jun 07, 2018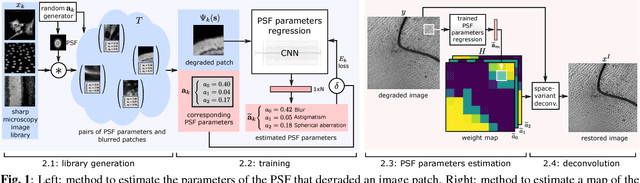
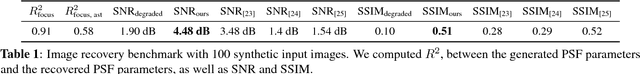
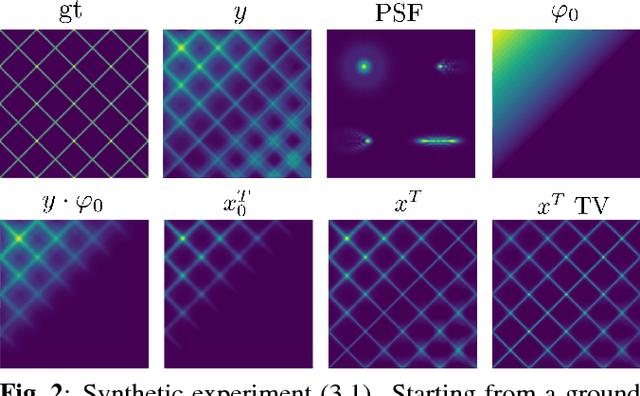
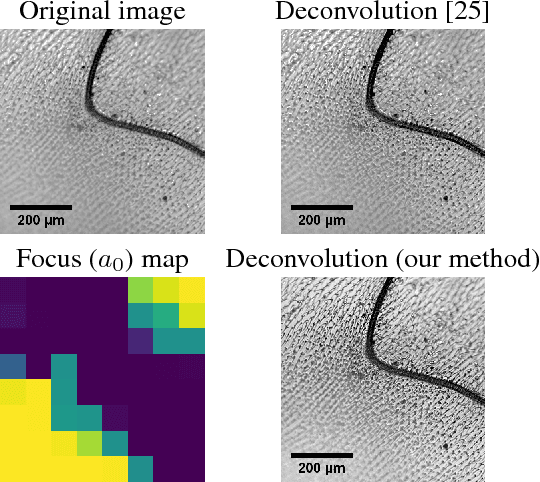
Abstract:We present a semi-blind, spatially-variant deconvolution technique aimed at optical microscopy that combines a local estimation step of the point spread function (PSF) and deconvolution using a spatially variant, regularized Richardson-Lucy algorithm. To find the local PSF map in a computationally tractable way, we train a convolutional neural network to perform regression of an optical parametric model on synthetically blurred image patches. We deconvolved both synthetic and experimentally-acquired data, and achieved an improvement of image SNR of 1.00 dB on average, compared to other deconvolution algorithms.
* 2018/02/11: submitted to IEEE ICIP 2018 - 2018/05/04: accepted to IEEE ICIP 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge