Estibaliz Gómez-de-Mariscal
Roadmap on Deep Learning for Microscopy
Mar 07, 2023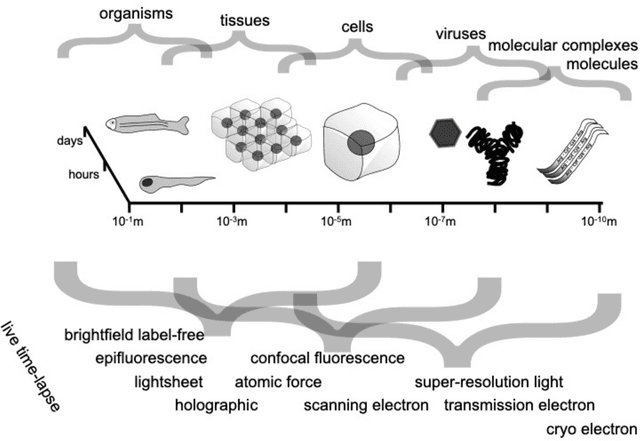
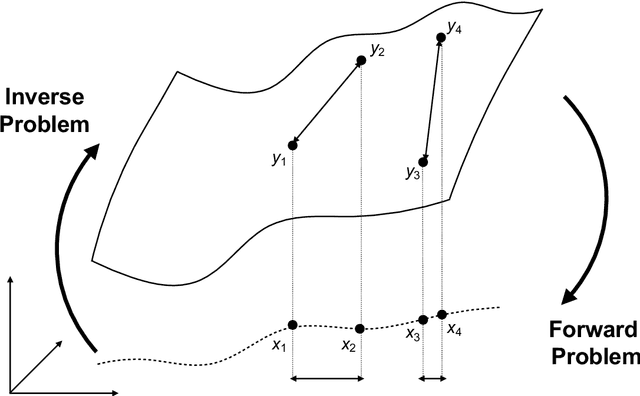
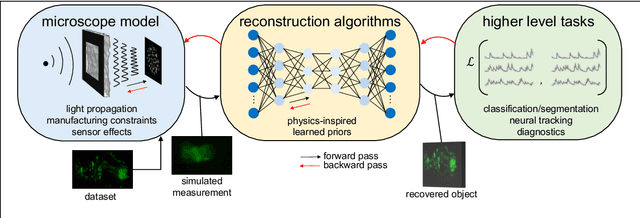
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Search for temporal cell segmentation robustness in phase-contrast microscopy videos
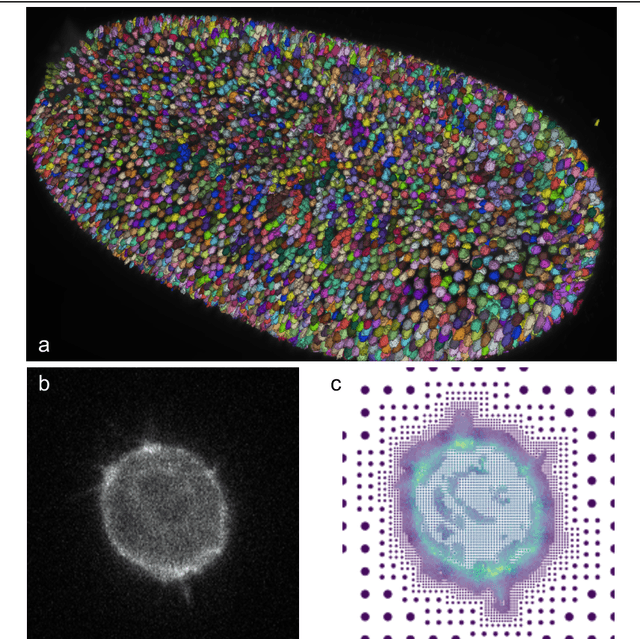
Dec 16, 2021Abstract:Studying cell morphology changes in time is critical to understanding cell migration mechanisms. In this work, we present a deep learning-based workflow to segment cancer cells embedded in 3D collagen matrices and imaged with phase-contrast microscopy. Our approach uses transfer learning and recurrent convolutional long-short term memory units to exploit the temporal information from the past and provide a consistent segmentation result. Lastly, we propose a geometrical-characterization approach to studying cancer cell morphology. Our approach provides stable results in time, and it is robust to the different weight initialization or training data sampling. We introduce a new annotated dataset for 2D cell segmentation and tracking, and an open-source implementation to replicate the experiments or adapt them to new image processing problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge