Daniel Franco-Barranco
Deep learning based domain adaptation for mitochondria segmentation on EM volumes
Feb 22, 2022



Abstract:Accurate segmentation of electron microscopy (EM) volumes of the brain is essential to characterize neuronal structures at a cell or organelle level. While supervised deep learning methods have led to major breakthroughs in that direction during the past years, they usually require large amounts of annotated data to be trained, and perform poorly on other data acquired under similar experimental and imaging conditions. This is a problem known as domain adaptation, since models that learned from a sample distribution (or source domain) struggle to maintain their performance on samples extracted from a different distribution or target domain. In this work, we address the complex case of deep learning based domain adaptation for mitochondria segmentation across EM datasets from different tissues and species. We present three unsupervised domain adaptation strategies to improve mitochondria segmentation in the target domain based on (1) state-of-the-art style transfer between images of both domains; (2) self-supervised learning to pre-train a model using unlabeled source and target images, and then fine-tune it only with the source labels; and (3) multi-task neural network architectures trained end-to-end with both labeled and unlabeled images. Additionally, we propose a new training stopping criterion based on morphological priors obtained exclusively in the source domain. We carried out all possible cross-dataset experiments using three publicly available EM datasets. We evaluated our proposed strategies on the mitochondria semantic labels predicted on the target datasets. The methods introduced here outperform the baseline methods and compare favorably to the state of the art. In the absence of validation labels, monitoring our proposed morphology-based metric is an intuitive and effective way to stop the training process and select in average optimal models.
Stable deep neural network architectures for mitochondria segmentation on electron microscopy volumes
Apr 08, 2021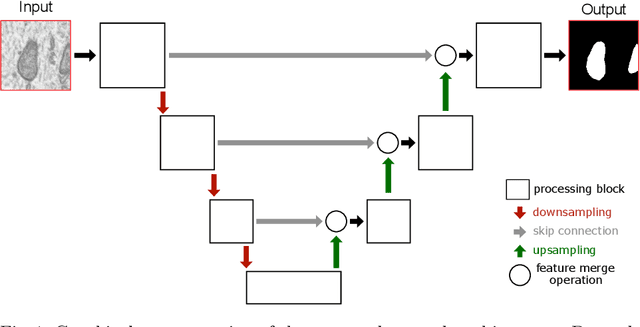
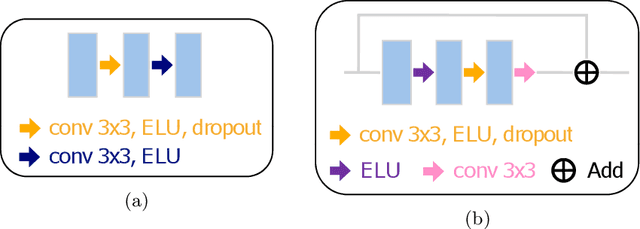
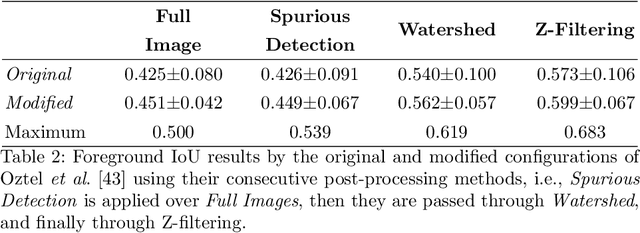
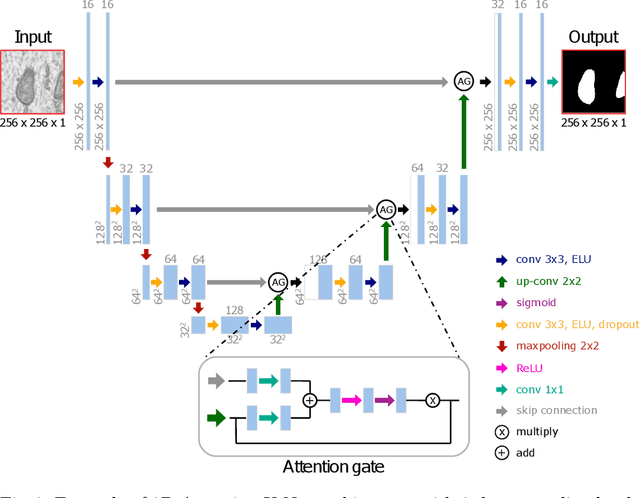
Abstract:Electron microscopy (EM) allows the identification of intracellular organelles such as mitochondria, providing insights for clinical and scientific studies. In recent years, a number of novel deep learning architectures have been published reporting superior performance, or even human-level accuracy, compared to previous approaches on public mitochondria segmentation datasets. Unfortunately, many of these publications do not make neither the code nor the full training details public to support the results obtained, leading to reproducibility issues and dubious model comparisons. For that reason, and following a recent code of best practices for reporting experimental results, we present an extensive study of the state-of-the-art deep learning architectures for the segmentation of mitochondria on EM volumes, and evaluate the impact in performance of different variations of 2D and 3D U-Net-like models for this task. To better understand the contribution of each component, a common set of pre- and post-processing operations has been implemented and tested with each approach. Moreover, an exhaustive sweep of hyperparameters values for all architectures have been performed and each configuration has been run multiple times to report the mean and standard deviation values of the evaluation metrics. Using this methodology, we found very stable architectures and hyperparameter configurations that consistently obtain state-of-the-art results in the well-known EPFL Hippocampus mitochondria segmentation dataset. Furthermore, we have benchmarked our proposed models on two other available datasets, Lucchi++ and Kasthuri++, where they outperform all previous works. The code derived from this research and its documentation are publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge