Daniel Sage
JDLL: A library to run Deep Learning models on Java bioimage informatics platforms
Jun 07, 2023Abstract:We present JDLL, an agile Java library that offers a comprehensive toolset/API to unify the development of high-end applications of DL for bioimage analysis and to streamline their installation and maintenance. JDLL provides all the functions required to consume DL models seamlessly, without being burdened by the configuration of the Python-based DL frameworks, within Java bioimage informatics platforms. Moreover, it allows the deployment of pre-trained models in the Bioimage Model Zoo (BMZ) by shipping the logic to connect to the BMZ website, download and run a selected model inference.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023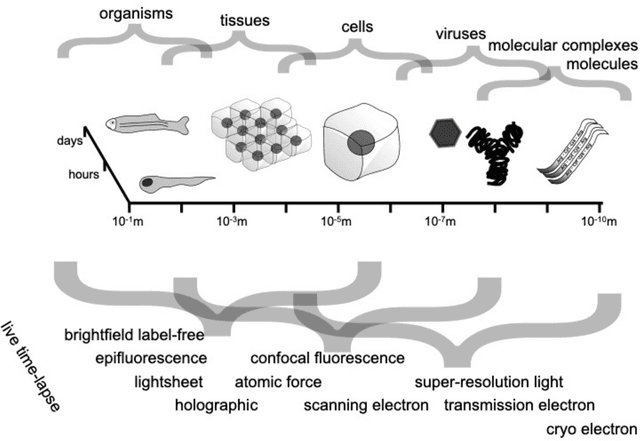
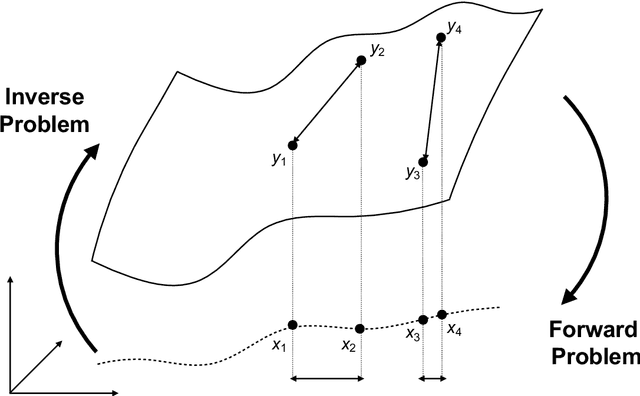
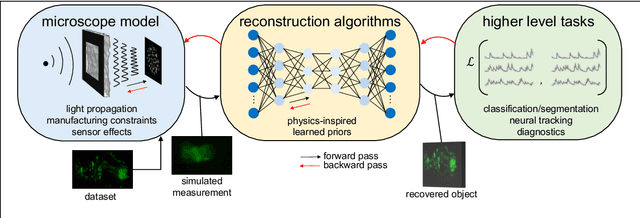
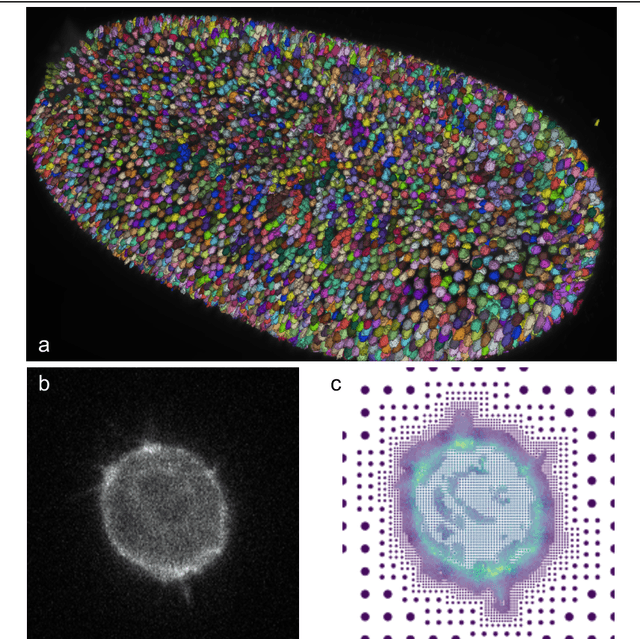
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Optimal transport-based metric for SMLM
Oct 26, 2020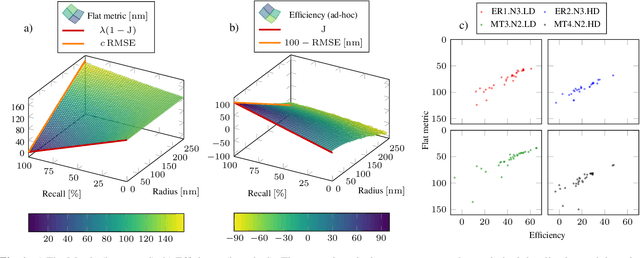
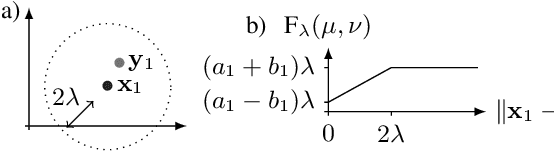
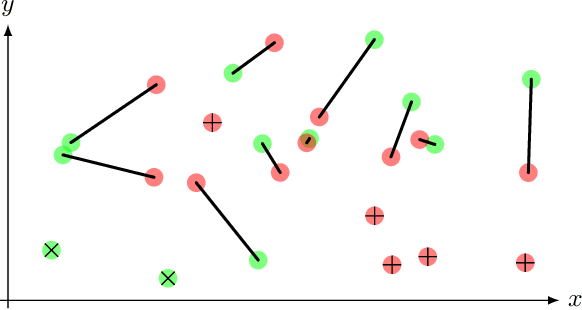
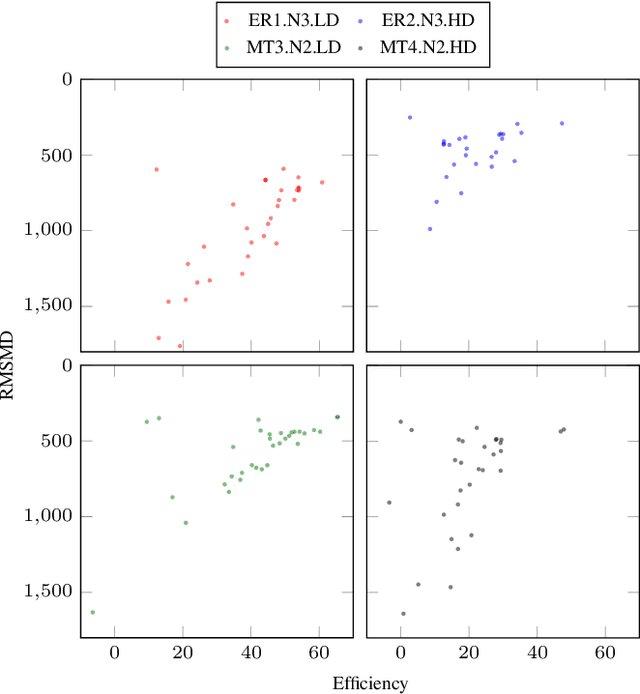
Abstract:We propose the use of Flat Metric to assess the performance of reconstruction methods for single-molecule localization microscopy (SMLM)in scenarios where the ground-truth is available. Flat Metric is intimately related to the concept of optimal transport between measures of different mass, providing solid mathematical foundations for SMLM evaluation and integrating both localization and detection performance. In this paper, we introduce the foundations of Flat Metric and validate this measure by applying it to controlled synthetic examples and to data from the SMLM 2016 Challenge.
W2S: A Joint Denoising and Super-Resolution Dataset
Mar 12, 2020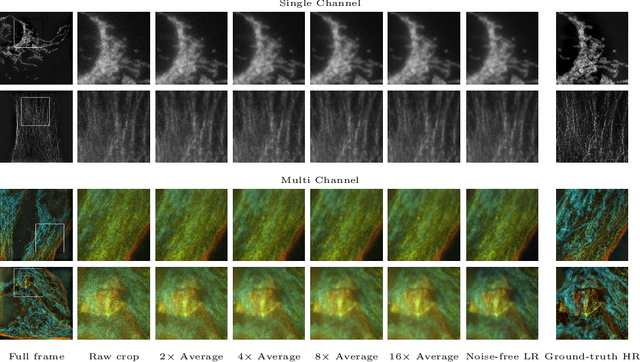
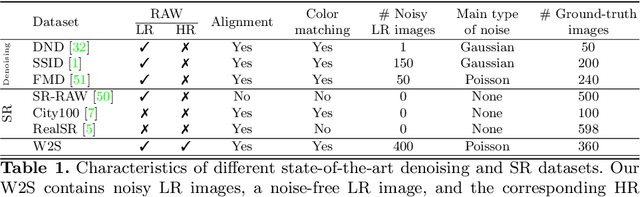
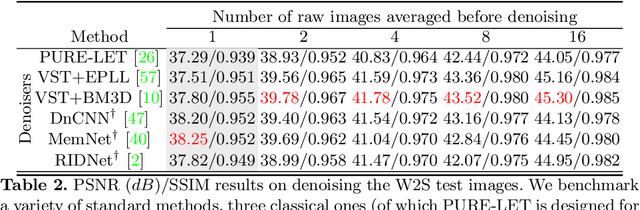
Abstract:Denoising and super-resolution (SR) are fundamental tasks in imaging. These two restoration tasks are well covered in the literature, however, only separately. Given a noisy low-resolution (LR) input image, it is yet unclear what the best approach would be in order to obtain a noise-free high-resolution (HR) image. In order to study joint denoising and super-resolution (JDSR), a dataset containing pairs of noisy LR images and the corresponding HR images is fundamental. We propose such a novel JDSR dataset, Wieldfield2SIM (W2S), acquired using microscopy equipment and techniques. W2S is comprised of 144,000 real fluorescence microscopy images, used to form a total of 360 sets of images. A set is comprised of noisy LR images with different noise levels, a noise-free LR image, and a corresponding high-quality HR image. W2S allows us to benchmark the combinations of 6 denoising methods and 6 SR methods. We show that state-of-the-art SR networks perform very poorly on noisy inputs, with a loss reaching 14dB relative to noise-free inputs. Our evaluation also shows that applying the best denoiser in terms of reconstruction error followed by the best SR method does not yield the best result. The best denoising PSNR can, for instance, come at the expense of a loss in high frequencies, which is detrimental for SR methods. We lastly demonstrate that a light-weight SR network with a novel texture loss, trained specifically for JDSR, outperforms any combination of state-of-the-art deep denoising and SR networks.
On The Continuous Steering of the Scale of Tight Wavelet Frames
Dec 07, 2015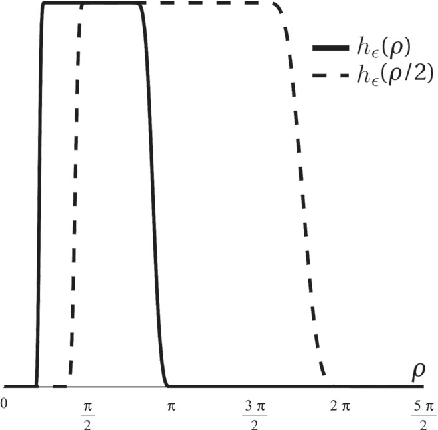
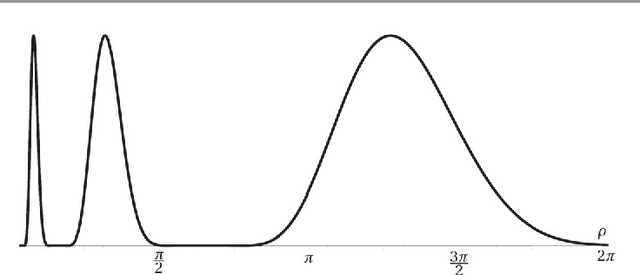
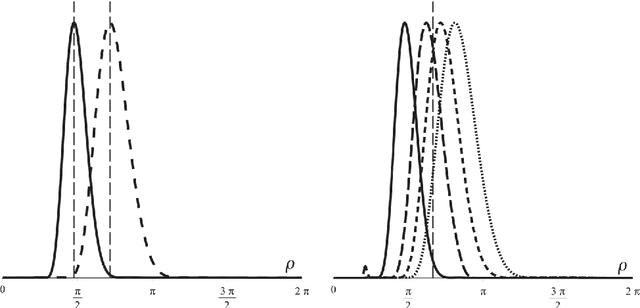
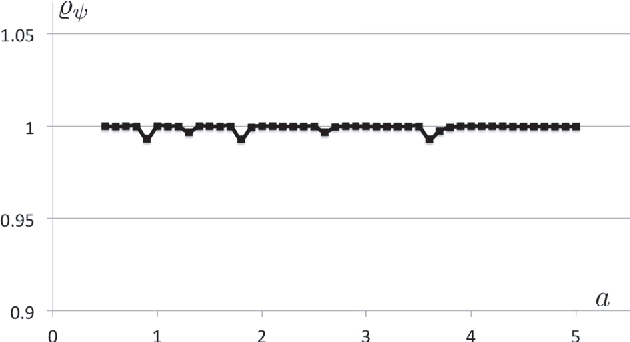
Abstract:In analogy with steerable wavelets, we present a general construction of adaptable tight wavelet frames, with an emphasis on scaling operations. In particular, the derived wavelets can be "dilated" by a procedure comparable to the operation of steering steerable wavelets. The fundamental aspects of the construction are the same: an admissible collection of Fourier multipliers is used to extend a tight wavelet frame, and the "scale" of the wavelets is adapted by scaling the multipliers. As an application, the proposed wavelets can be used to improve the frequency localization. Importantly, the localized frequency bands specified by this construction can be scaled efficiently using matrix multiplication.
Fast O(1) bilateral filtering using trigonometric range kernels
Jul 27, 2011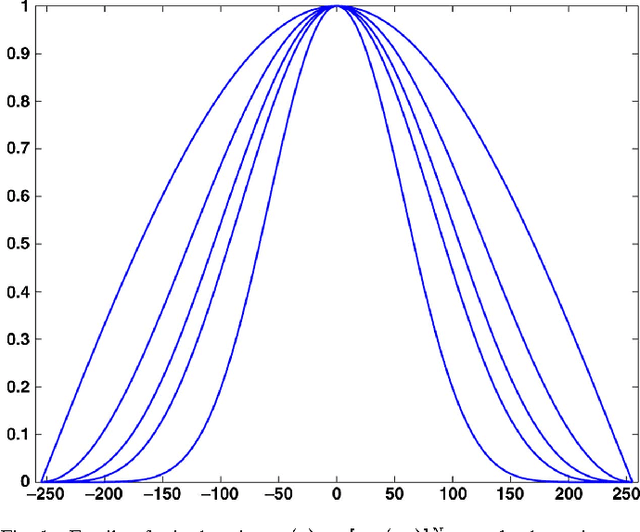
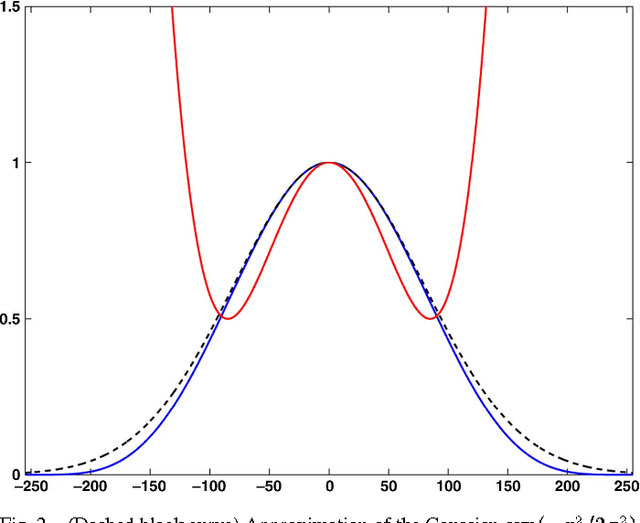
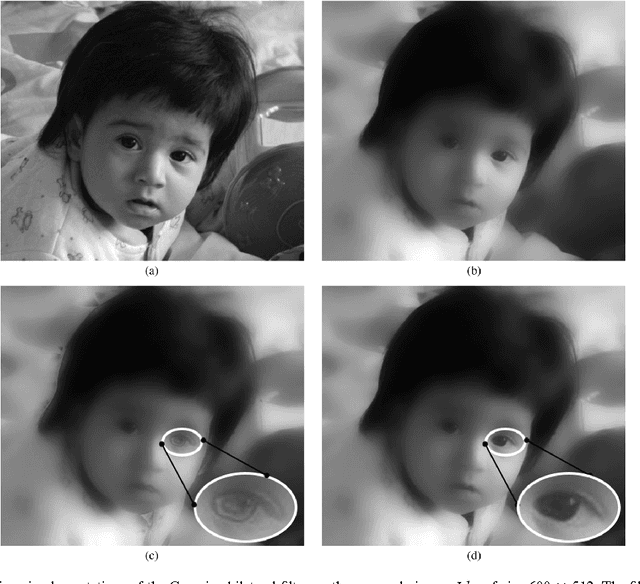

Abstract:It is well-known that spatial averaging can be realized (in space or frequency domain) using algorithms whose complexity does not depend on the size or shape of the filter. These fast algorithms are generally referred to as constant-time or O(1) algorithms in the image processing literature. Along with the spatial filter, the edge-preserving bilateral filter [Tomasi1998] involves an additional range kernel. This is used to restrict the averaging to those neighborhood pixels whose intensity are similar or close to that of the pixel of interest. The range kernel operates by acting on the pixel intensities. This makes the averaging process non-linear and computationally intensive, especially when the spatial filter is large. In this paper, we show how the O(1) averaging algorithms can be leveraged for realizing the bilateral filter in constant-time, by using trigonometric range kernels. This is done by generalizing the idea in [Porikli2008] of using polynomial range kernels. The class of trigonometric kernels turns out to be sufficiently rich, allowing for the approximation of the standard Gaussian bilateral filter. The attractive feature of our approach is that, for a fixed number of terms, the quality of approximation achieved using trigonometric kernels is much superior to that obtained in [Porikli2008] using polynomials.
* Accepted in IEEE Transactions on Image Processing. Also see addendum: https://sites.google.com/site/kunalspage/home/Addendum.pdf
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge