Indika Perera
Parallel Algorithms for Densest Subgraph Discovery Using Shared Memory Model
Feb 27, 2021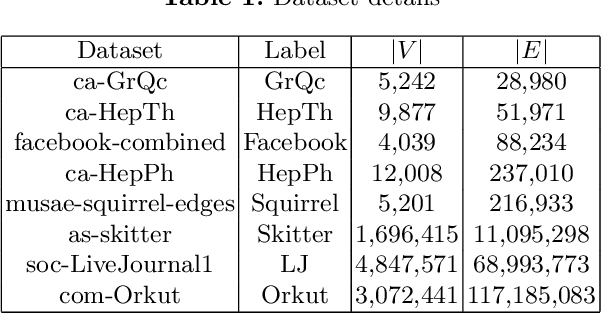
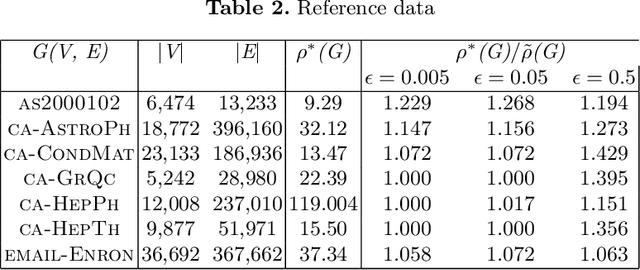
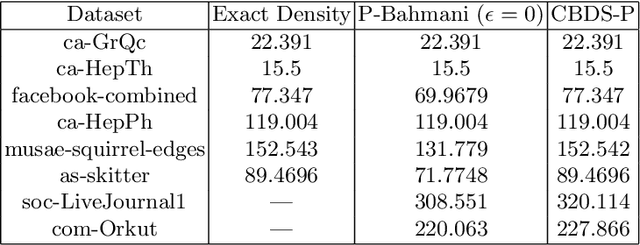
Abstract:The problem of finding dense components of a graph is a widely explored area in data analysis, with diverse applications in fields and branches of study including community mining, spam detection, computer security and bioinformatics. This research project explores previously available algorithms in order to study them and identify potential modifications that could result in an improved version with considerable performance and efficiency leap. Furthermore, efforts were also steered towards devising a novel algorithm for the problem of densest subgraph discovery. This paper presents an improved implementation of a widely used densest subgraph discovery algorithm and a novel parallel algorithm which produces better results than a 2-approximation.
Survival prediction and risk estimation of Glioma patients using mRNA expressions
Nov 02, 2020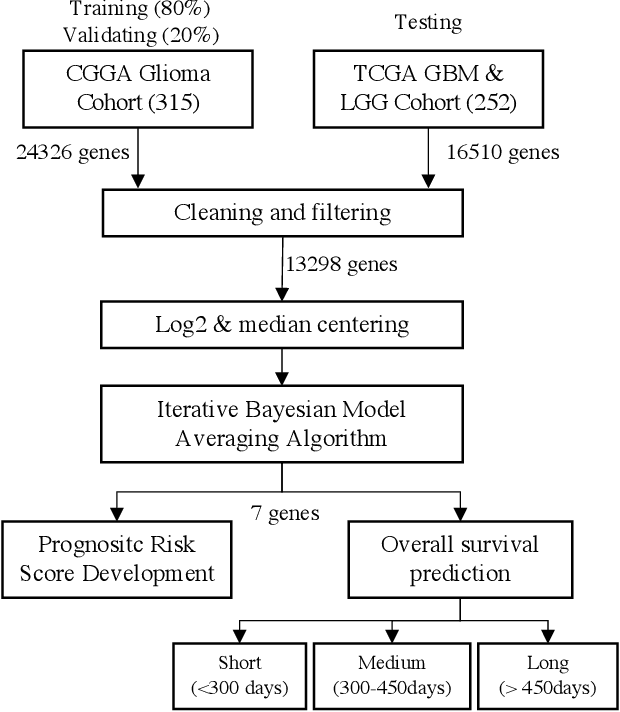
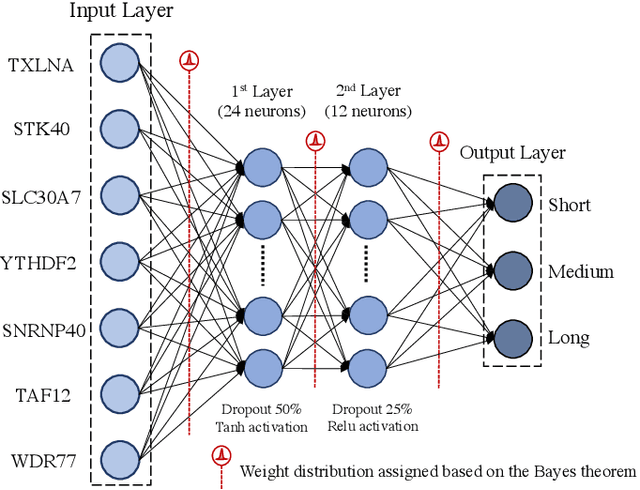
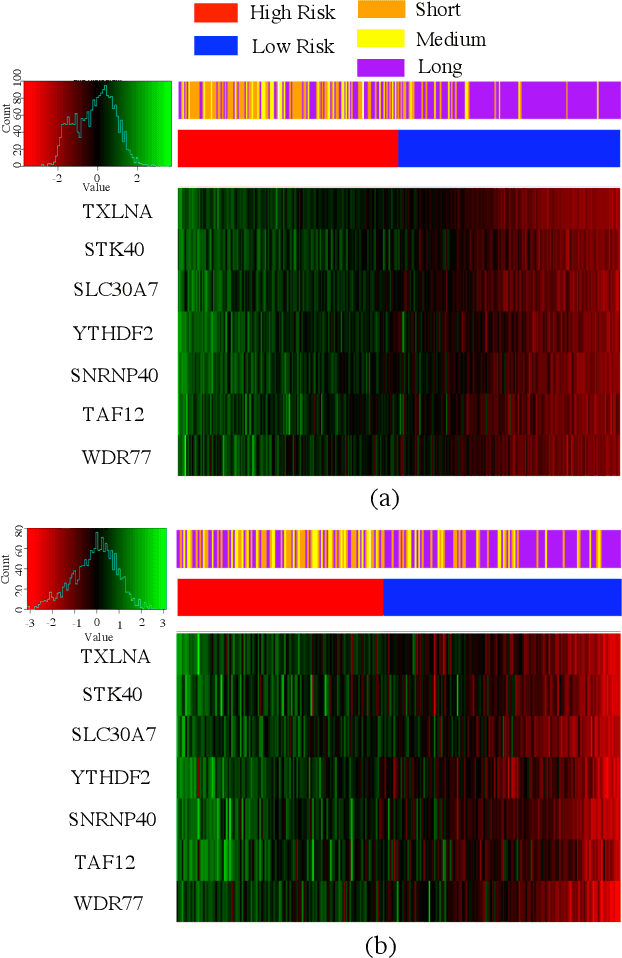
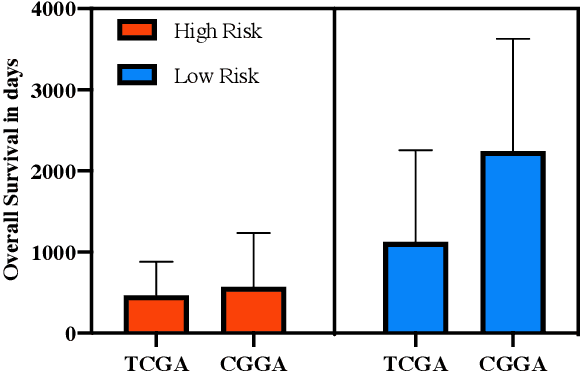
Abstract:Gliomas are lethal type of central nervous system tumors with a poor prognosis. Recently, with the advancements in the micro-array technologies thousands of gene expression related data of glioma patients are acquired, leading for salient analysis in many aspects. Thus, genomics are been emerged into the field of prognosis analysis. In this work, we identify survival related 7 gene signature and explore two approaches for survival prediction and risk estimation. For survival prediction, we propose a novel probabilistic programming based approach, which outperforms the existing traditional machine learning algorithms. An average 4 fold accuracy of 74% is obtained with the proposed algorithm. Further, we construct a prognostic risk model for risk estimation of glioma patients. This model reflects the survival of glioma patients, with high risk for low survival patients.
Radiogenomics of Glioblastoma: Identification of Radiomics associated with Molecular Subtypes
Oct 27, 2020
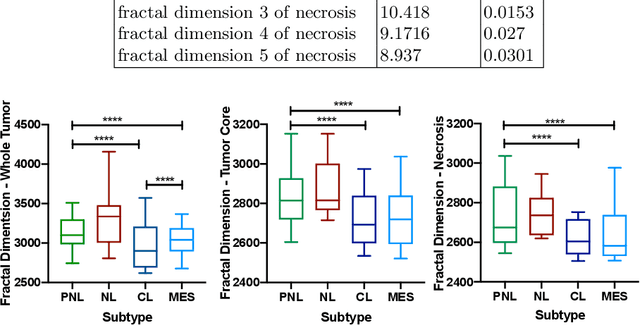
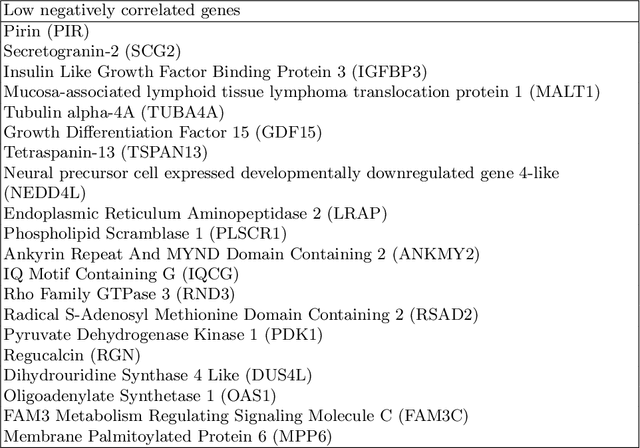
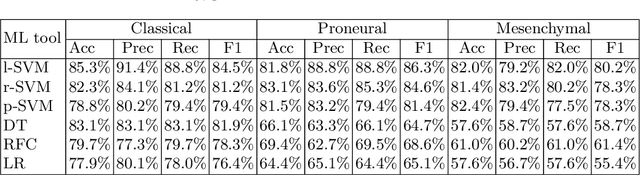
Abstract:Glioblastoma is the most malignant type of central nervous system tumor with GBM subtypes cleaved based on molecular level gene alterations. These alterations are also happened to affect the histology. Thus, it can cause visible changes in images, such as enhancement and edema development. In this study, we extract intensity, volume, and texture features from the tumor subregions to identify the correlations with gene expression features and overall survival. Consequently, we utilize the radiomics to find associations with the subtypes of glioblastoma. Accordingly, the fractal dimensions of the whole tumor, tumor core, and necrosis regions show a significant difference between the Proneural, Classical and Mesenchymal subtypes. Additionally, the subtypes of GBM are predicted with an average accuracy of 79% utilizing radiomics and accuracy over 90% utilizing gene expression profiles.
Cognitive Analysis of 360 degree Surround Photos
Jan 17, 2019
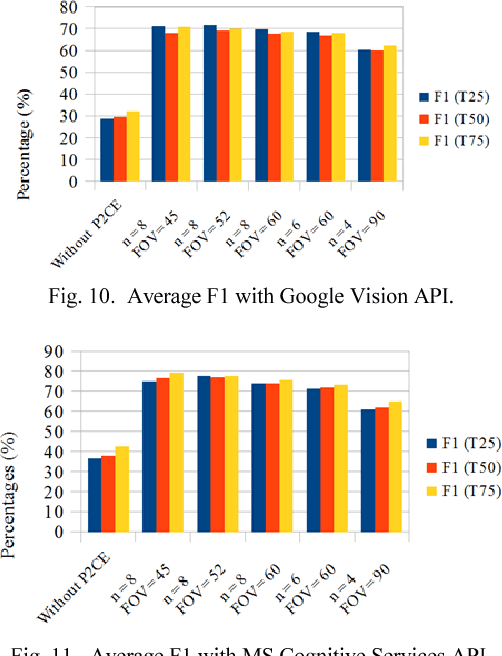
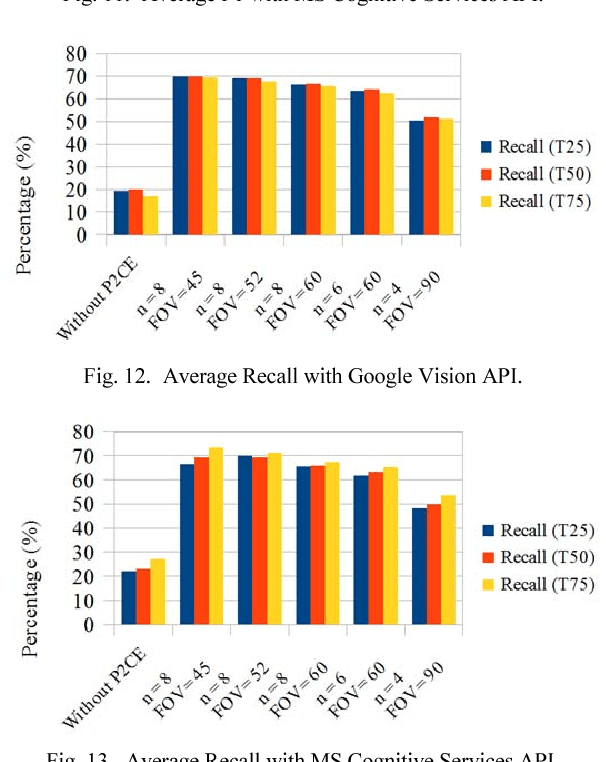
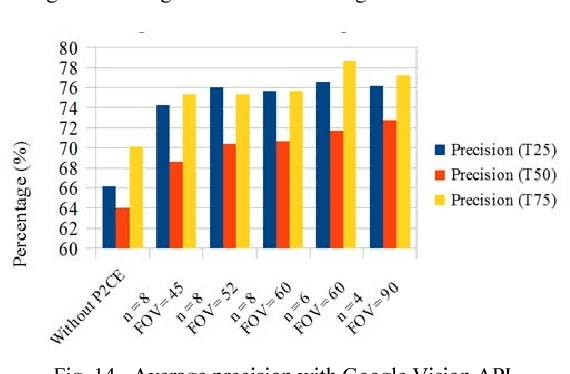
Abstract:360 degrees surround photography or photospheres have taken the world by storm as the new media for content creation providing viewers rich, immersive experience compared to conventional photography. With the emergence of Virtual Reality as a mainstream trend, the 360 degrees photography is increasingly important to offer a practical approach to the general public to capture virtual reality ready content from their mobile phones without explicit tool support or knowledge. Even though the amount of 360-degree surround content being uploaded to the Internet continues to grow, there is no proper way to index them or to process them for further information. This is because of the difficulty in image processing the photospheres due to the distorted nature of objects embedded. This challenge lies in the way 360-degree panoramic photospheres are saved. This paper presents a unique, and innovative technique named Photosphere to Cognition Engine (P2CE), which allows cognitive analysis on 360-degree surround photos using existing image cognitive analysis algorithms and APIs designed for conventional photos. We have optimized the system using a wide variety of indoor and outdoor samples and extensive evaluation approaches. On average, P2CE provides up-to 100% growth in accuracy on image cognitive analysis of Photospheres over direct use of conventional non-photosphere based Image Cognition Systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge