H. Rolf Jäger
Generalizable automated ischaemic stroke lesion segmentation with vision transformers
Feb 10, 2025
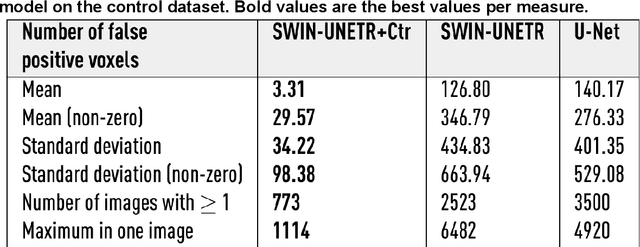
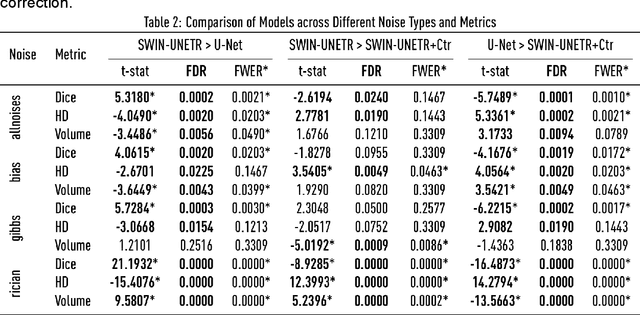
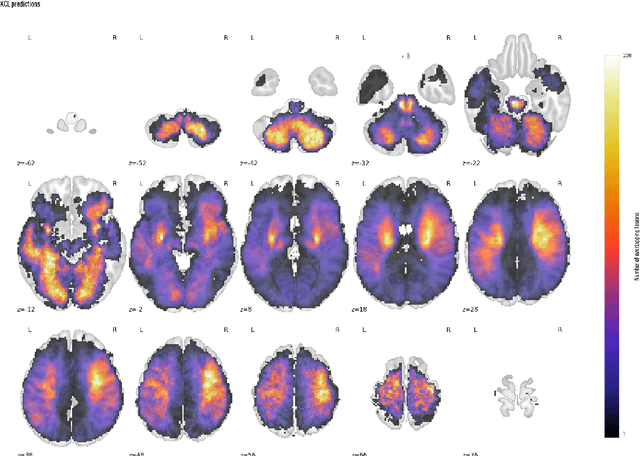
Abstract:Ischaemic stroke, a leading cause of death and disability, critically relies on neuroimaging for characterising the anatomical pattern of injury. Diffusion-weighted imaging (DWI) provides the highest expressivity in ischemic stroke but poses substantial challenges for automated lesion segmentation: susceptibility artefacts, morphological heterogeneity, age-related comorbidities, time-dependent signal dynamics, instrumental variability, and limited labelled data. Current U-Net-based models therefore underperform, a problem accentuated by inadequate evaluation metrics that focus on mean performance, neglecting anatomical, subpopulation, and acquisition-dependent variability. Here, we present a high-performance DWI lesion segmentation tool addressing these challenges through optimized vision transformer-based architectures, integration of 3563 annotated lesions from multi-site data, and algorithmic enhancements, achieving state-of-the-art results. We further propose a novel evaluative framework assessing model fidelity, equity (across demographics and lesion subtypes), anatomical precision, and robustness to instrumental variability, promoting clinical and research utility. This work advances stroke imaging by reconciling model expressivity with domain-specific challenges and redefining performance benchmarks to prioritize equity and generalizability, critical for personalized medicine and mechanistic research.
Unsupervised 3D out-of-distribution detection with latent diffusion models
Jul 07, 2023


Abstract:Methods for out-of-distribution (OOD) detection that scale to 3D data are crucial components of any real-world clinical deep learning system. Classic denoising diffusion probabilistic models (DDPMs) have been recently proposed as a robust way to perform reconstruction-based OOD detection on 2D datasets, but do not trivially scale to 3D data. In this work, we propose to use Latent Diffusion Models (LDMs), which enable the scaling of DDPMs to high-resolution 3D medical data. We validate the proposed approach on near- and far-OOD datasets and compare it to a recently proposed, 3D-enabled approach using Latent Transformer Models (LTMs). Not only does the proposed LDM-based approach achieve statistically significant better performance, it also shows less sensitivity to the underlying latent representation, more favourable memory scaling, and produces better spatial anomaly maps. Code is available at https://github.com/marksgraham/ddpm-ood
3D multirater RCNN for multimodal multiclass detection and characterisation of extremely small objects
Dec 21, 2018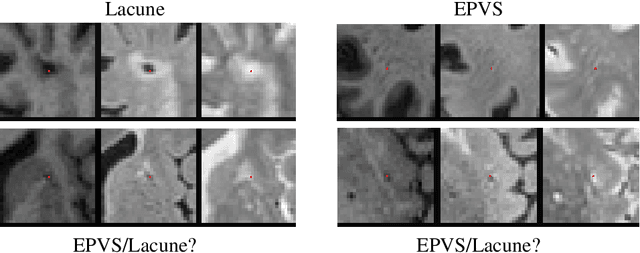
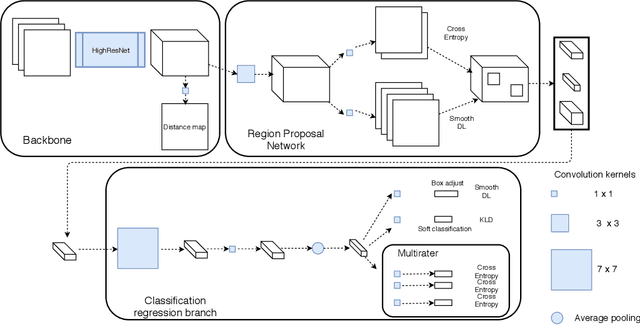
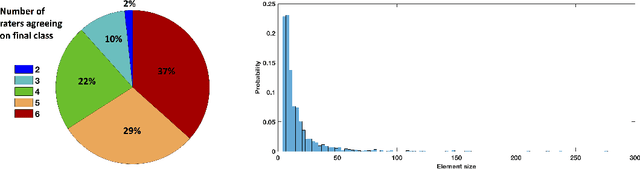
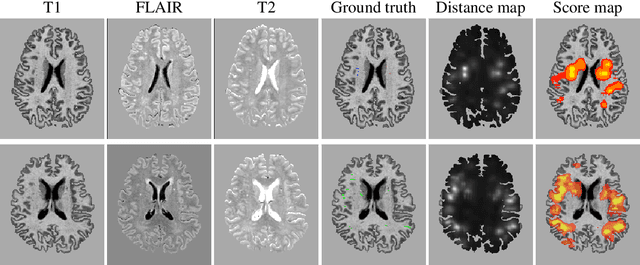
Abstract:Extremely small objects (ESO) have become observable on clinical routine magnetic resonance imaging acquisitions, thanks to a reduction in acquisition time at higher resolution. Despite their small size (usually $<$10 voxels per object for an image of more than $10^6$ voxels), these markers reflect tissue damage and need to be accounted for to investigate the complete phenotype of complex pathological pathways. In addition to their very small size, variability in shape and appearance leads to high labelling variability across human raters, resulting in a very noisy gold standard. Such objects are notably present in the context of cerebral small vessel disease where enlarged perivascular spaces and lacunes, commonly observed in the ageing population, are thought to be associated with acceleration of cognitive decline and risk of dementia onset. In this work, we redesign the RCNN model to scale to 3D data, and to jointly detect and characterise these important markers of age-related neurovascular changes. We also propose training strategies enforcing the detection of extremely small objects, ensuring a tractable and stable training process.
VTrails: Inferring Vessels with Geodesic Connectivity Trees
Jun 08, 2018



Abstract:The analysis of vessel morphology and connectivity has an impact on a number of cardiovascular and neurovascular applications by providing patient-specific high-level quantitative features such as spatial location, direction and scale. In this paper we present an end-to-end approach to extract an acyclic vascular tree from angiographic data by solving a connectivity-enforcing anisotropic fast marching over a voxel-wise tensor field representing the orientation of the underlying vascular tree. The method is validated using synthetic and real vascular images. We compare VTrails against classical and state-of-the-art ridge detectors for tubular structures by assessing the connectedness of the vesselness map and inspecting the synthesized tensor field as proof of concept. VTrails performance is evaluated on images with different levels of degradation: we verify that the extracted vascular network is an acyclic graph (i.e. a tree), and we report the extraction accuracy, precision and recall.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge