David Werring
Unsupervised 3D out-of-distribution detection with latent diffusion models
Jul 07, 2023


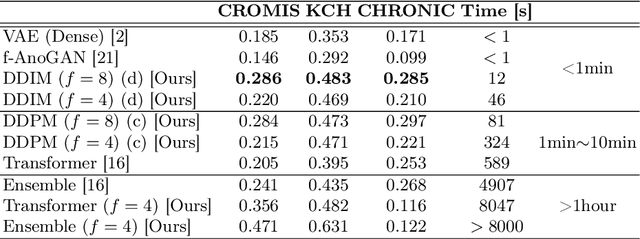
Abstract:Methods for out-of-distribution (OOD) detection that scale to 3D data are crucial components of any real-world clinical deep learning system. Classic denoising diffusion probabilistic models (DDPMs) have been recently proposed as a robust way to perform reconstruction-based OOD detection on 2D datasets, but do not trivially scale to 3D data. In this work, we propose to use Latent Diffusion Models (LDMs), which enable the scaling of DDPMs to high-resolution 3D medical data. We validate the proposed approach on near- and far-OOD datasets and compare it to a recently proposed, 3D-enabled approach using Latent Transformer Models (LTMs). Not only does the proposed LDM-based approach achieve statistically significant better performance, it also shows less sensitivity to the underlying latent representation, more favourable memory scaling, and produces better spatial anomaly maps. Code is available at https://github.com/marksgraham/ddpm-ood
Fast Unsupervised Brain Anomaly Detection and Segmentation with Diffusion Models
Jun 07, 2022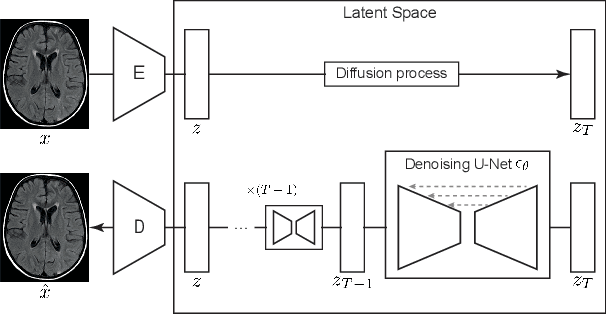
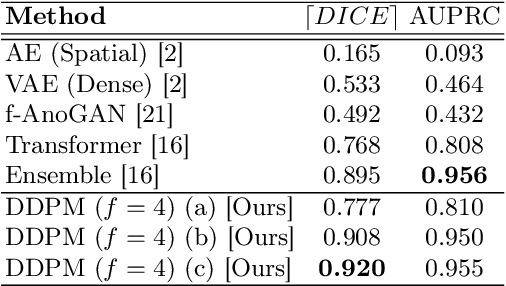
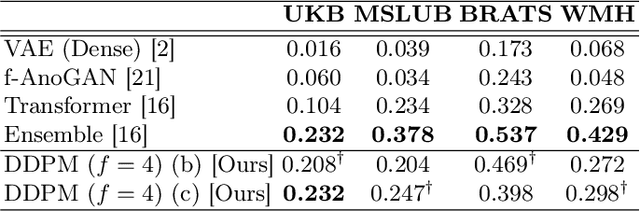
Abstract:Deep generative models have emerged as promising tools for detecting arbitrary anomalies in data, dispensing with the necessity for manual labelling. Recently, autoregressive transformers have achieved state-of-the-art performance for anomaly detection in medical imaging. Nonetheless, these models still have some intrinsic weaknesses, such as requiring images to be modelled as 1D sequences, the accumulation of errors during the sampling process, and the significant inference times associated with transformers. Denoising diffusion probabilistic models are a class of non-autoregressive generative models recently shown to produce excellent samples in computer vision (surpassing Generative Adversarial Networks), and to achieve log-likelihoods that are competitive with transformers while having fast inference times. Diffusion models can be applied to the latent representations learnt by autoencoders, making them easily scalable and great candidates for application to high dimensional data, such as medical images. Here, we propose a method based on diffusion models to detect and segment anomalies in brain imaging. By training the models on healthy data and then exploring its diffusion and reverse steps across its Markov chain, we can identify anomalous areas in the latent space and hence identify anomalies in the pixel space. Our diffusion models achieve competitive performance compared with autoregressive approaches across a series of experiments with 2D CT and MRI data involving synthetic and real pathological lesions with much reduced inference times, making their usage clinically viable.
Transformer-based out-of-distribution detection for clinically safe segmentation
May 21, 2022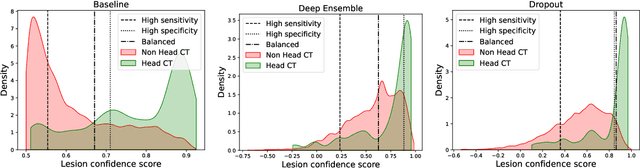
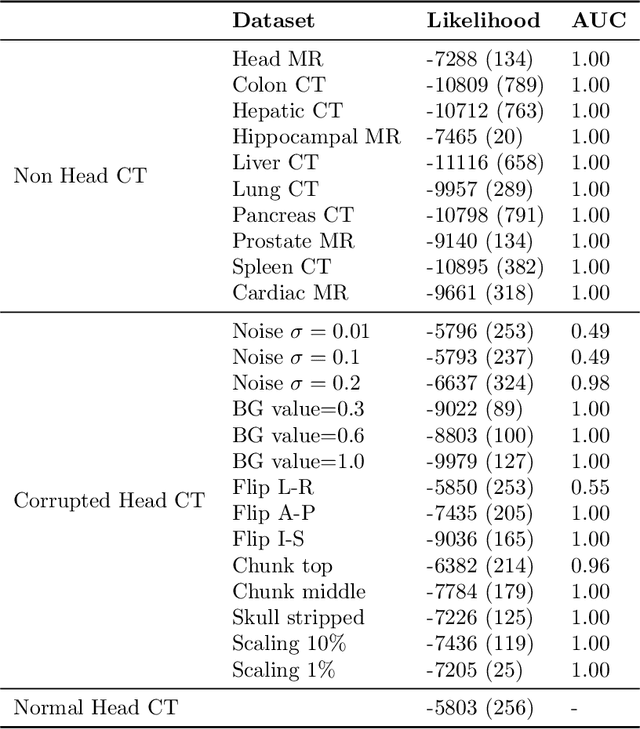
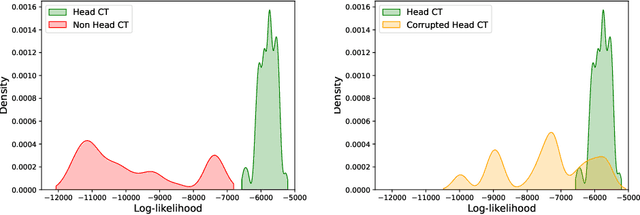
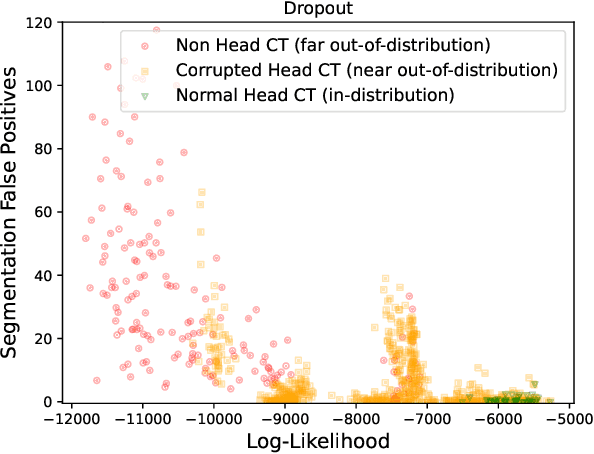
Abstract:In a clinical setting it is essential that deployed image processing systems are robust to the full range of inputs they might encounter and, in particular, do not make confidently wrong predictions. The most popular approach to safe processing is to train networks that can provide a measure of their uncertainty, but these tend to fail for inputs that are far outside the training data distribution. Recently, generative modelling approaches have been proposed as an alternative; these can quantify the likelihood of a data sample explicitly, filtering out any out-of-distribution (OOD) samples before further processing is performed. In this work, we focus on image segmentation and evaluate several approaches to network uncertainty in the far-OOD and near-OOD cases for the task of segmenting haemorrhages in head CTs. We find all of these approaches are unsuitable for safe segmentation as they provide confidently wrong predictions when operating OOD. We propose performing full 3D OOD detection using a VQ-GAN to provide a compressed latent representation of the image and a transformer to estimate the data likelihood. Our approach successfully identifies images in both the far- and near-OOD cases. We find a strong relationship between image likelihood and the quality of a model's segmentation, making this approach viable for filtering images unsuitable for segmentation. To our knowledge, this is the first time transformers have been applied to perform OOD detection on 3D image data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge