James Teo
RelCAT: Advancing Extraction of Clinical Inter-Entity Relationships from Unstructured Electronic Health Records
Jan 27, 2025Abstract:This study introduces RelCAT (Relation Concept Annotation Toolkit), an interactive tool, library, and workflow designed to classify relations between entities extracted from clinical narratives. Building upon the CogStack MedCAT framework, RelCAT addresses the challenge of capturing complete clinical relations dispersed within text. The toolkit implements state-of-the-art machine learning models such as BERT and Llama along with proven evaluation and training methods. We demonstrate a dataset annotation tool (built within MedCATTrainer), model training, and evaluate our methodology on both openly available gold-standard and real-world UK National Health Service (NHS) hospital clinical datasets. We perform extensive experimentation and a comparative analysis of the various publicly available models with varied approaches selected for model fine-tuning. Finally, we achieve macro F1-scores of 0.977 on the gold-standard n2c2, surpassing the previous state-of-the-art performance, and achieve performance of >=0.93 F1 on our NHS gathered datasets.
Large Language Models for Medical Forecasting -- Foresight 2
Dec 14, 2024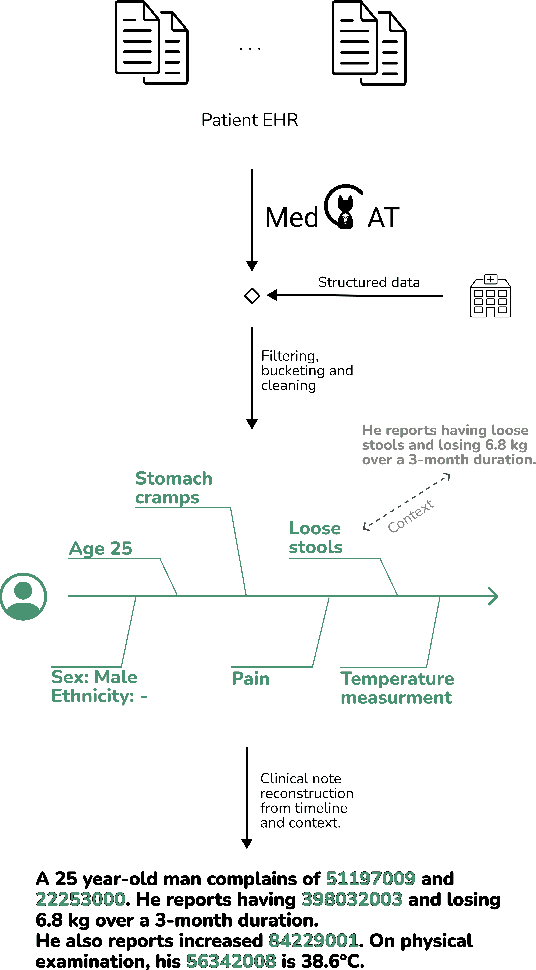
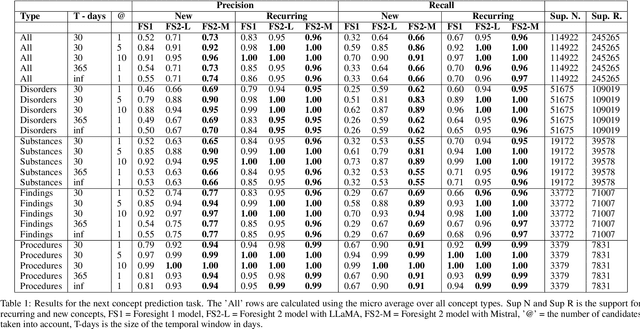
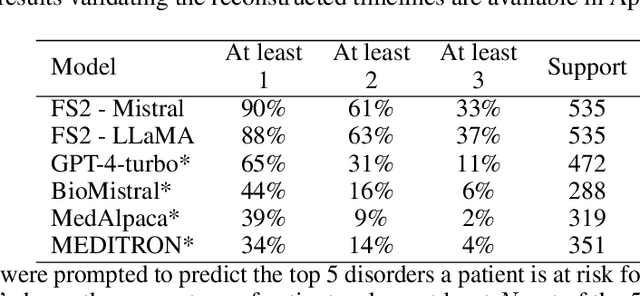
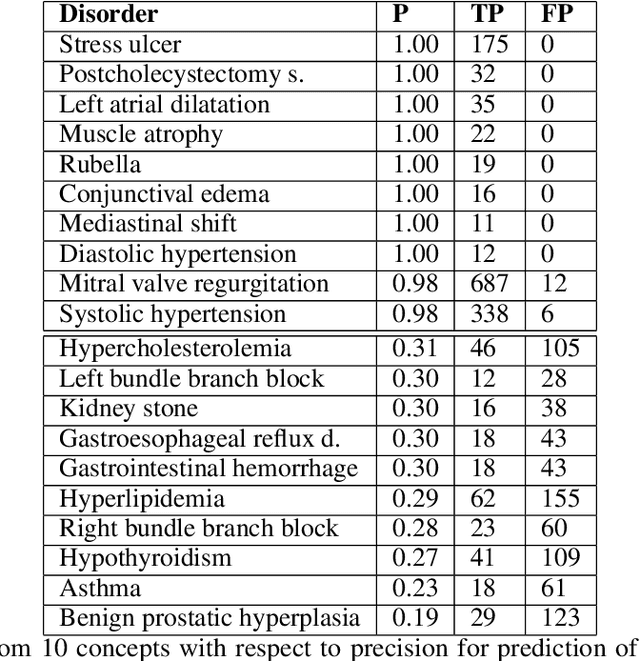
Abstract:Foresight 2 (FS2) is a large language model fine-tuned on hospital data for modelling patient timelines (GitHub 'removed for anon'). It can understand patients' clinical notes and predict SNOMED codes for a wide range of biomedical use cases, including diagnosis suggestions, risk forecasting, and procedure and medication recommendations. FS2 is trained on the free text portion of the MIMIC-III dataset, firstly through extracting biomedical concepts and then creating contextualised patient timelines, upon which the model is then fine-tuned. The results show significant improvement over the previous state-of-the-art for the next new biomedical concept prediction (P/R - 0.73/0.66 vs 0.52/0.32) and a similar improvement specifically for the next new disorder prediction (P/R - 0.69/0.62 vs 0.46/0.25). Finally, on the task of risk forecast, we compare our model to GPT-4-turbo (and a range of open-source biomedical LLMs) and show that FS2 performs significantly better on such tasks (P@5 - 0.90 vs 0.65). This highlights the need to incorporate hospital data into LLMs and shows that small models outperform much larger ones when fine-tuned on high-quality, specialised data.
Improving Extraction of Clinical Event Contextual Properties from Electronic Health Records: A Comparative Study
Aug 30, 2024
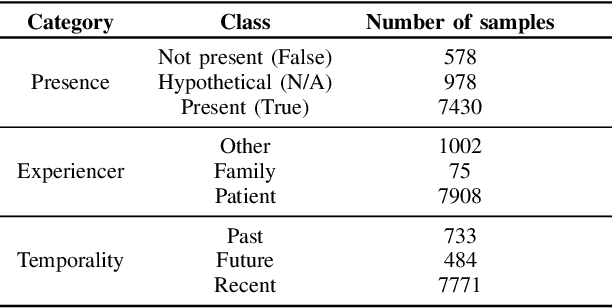
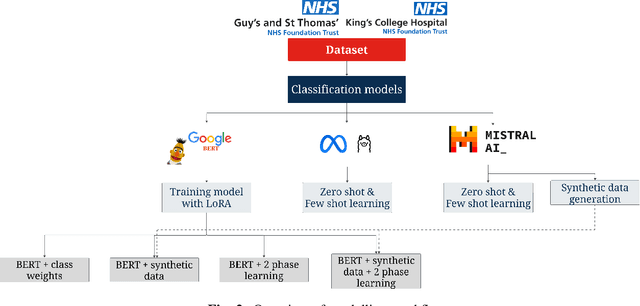
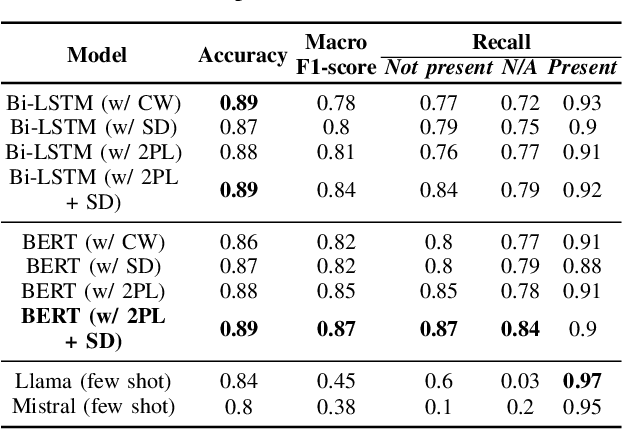
Abstract:Electronic Health Records are large repositories of valuable clinical data, with a significant portion stored in unstructured text format. This textual data includes clinical events (e.g., disorders, symptoms, findings, medications and procedures) in context that if extracted accurately at scale can unlock valuable downstream applications such as disease prediction. Using an existing Named Entity Recognition and Linking methodology, MedCAT, these identified concepts need to be further classified (contextualised) for their relevance to the patient, and their temporal and negated status for example, to be useful downstream. This study performs a comparative analysis of various natural language models for medical text classification. Extensive experimentation reveals the effectiveness of transformer-based language models, particularly BERT. When combined with class imbalance mitigation techniques, BERT outperforms Bi-LSTM models by up to 28% and the baseline BERT model by up to 16% for recall of the minority classes. The method has been implemented as part of CogStack/MedCAT framework and made available to the community for further research.
Framework to generate perfusion map from CT and CTA images in patients with acute ischemic stroke: A longitudinal and cross-sectional study
Apr 05, 2024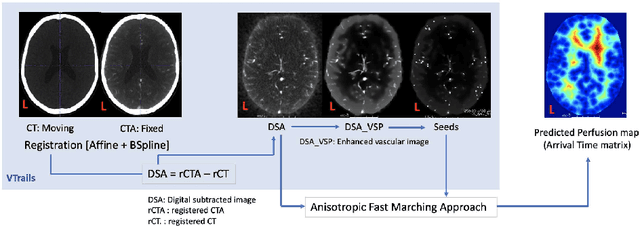
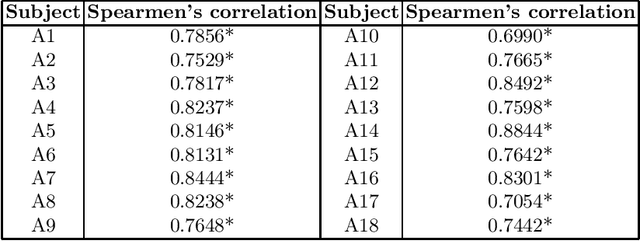
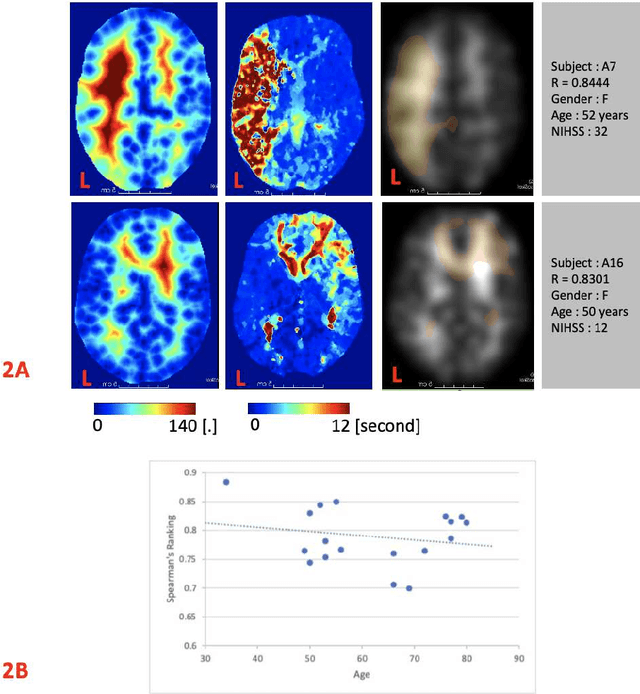
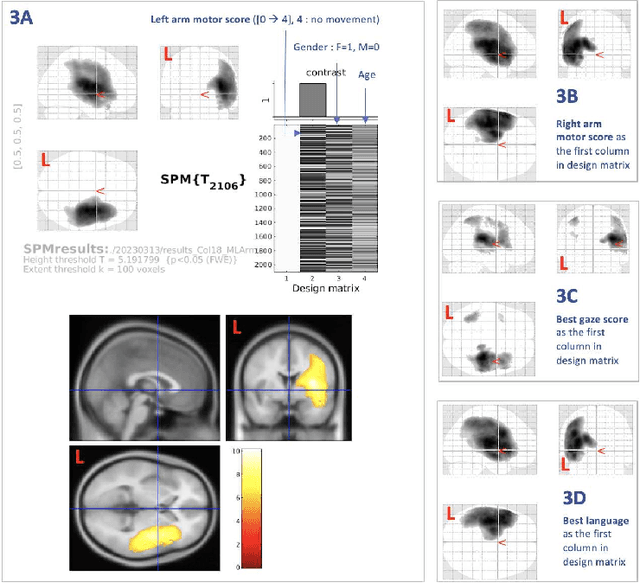
Abstract:Stroke is a leading cause of disability and death. Effective treatment decisions require early and informative vascular imaging. 4D perfusion imaging is ideal but rarely available within the first hour after stroke, whereas plain CT and CTA usually are. Hence, we propose a framework to extract a predicted perfusion map (PPM) derived from CT and CTA images. In all eighteen patients, we found significantly high spatial similarity (with average Spearman's correlation = 0.7893) between our predicted perfusion map (PPM) and the T-max map derived from 4D-CTP. Voxelwise correlations between the PPM and National Institutes of Health Stroke Scale (NIHSS) subscores for L/R hand motor, gaze, and language on a large cohort of 2,110 subjects reliably mapped symptoms to expected infarct locations. Therefore our PPM could serve as an alternative for 4D perfusion imaging, if the latter is unavailable, to investigate blood perfusion in the first hours after hospital admission.
Validating transformers for redaction of text from electronic health records in real-world healthcare
Oct 05, 2023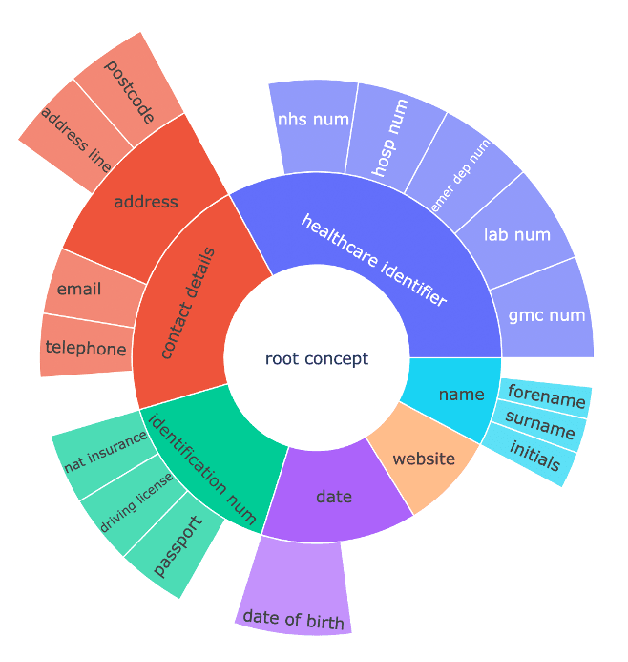
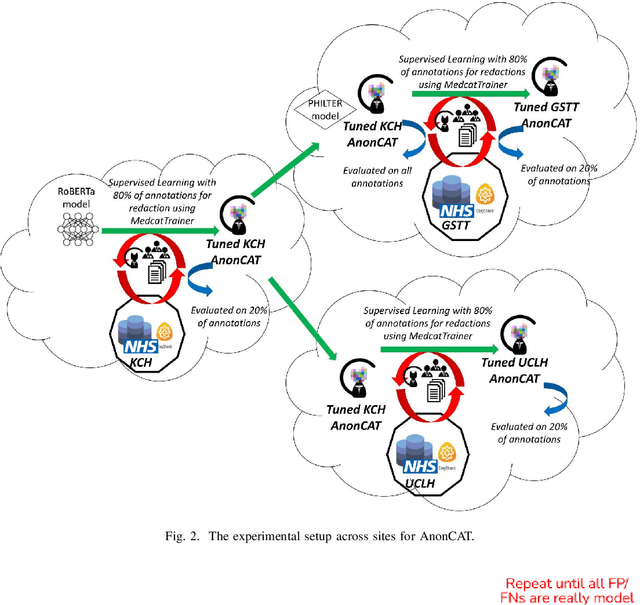
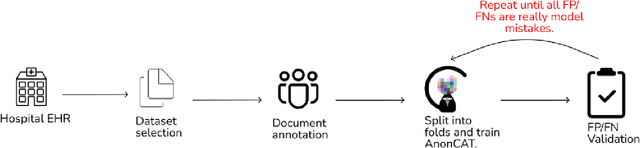
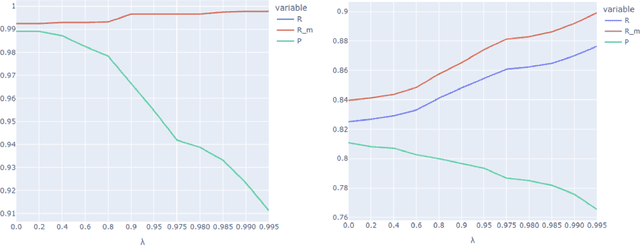
Abstract:Protecting patient privacy in healthcare records is a top priority, and redaction is a commonly used method for obscuring directly identifiable information in text. Rule-based methods have been widely used, but their precision is often low causing over-redaction of text and frequently not being adaptable enough for non-standardised or unconventional structures of personal health information. Deep learning techniques have emerged as a promising solution, but implementing them in real-world environments poses challenges due to the differences in patient record structure and language across different departments, hospitals, and countries. In this study, we present AnonCAT, a transformer-based model and a blueprint on how deidentification models can be deployed in real-world healthcare. AnonCAT was trained through a process involving manually annotated redactions of real-world documents from three UK hospitals with different electronic health record systems and 3116 documents. The model achieved high performance in all three hospitals with a Recall of 0.99, 0.99 and 0.96. Our findings demonstrate the potential of deep learning techniques for improving the efficiency and accuracy of redaction in global healthcare data and highlight the importance of building workflows which not just use these models but are also able to continually fine-tune and audit the performance of these algorithms to ensure continuing effectiveness in real-world settings. This approach provides a blueprint for the real-world use of de-identifying algorithms through fine-tuning and localisation, the code together with tutorials is available on GitHub (https://github.com/CogStack/MedCAT).
Summarisation of Electronic Health Records with Clinical Concept Guidance
Nov 14, 2022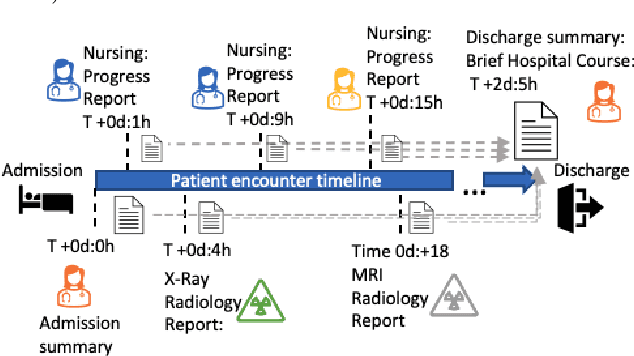
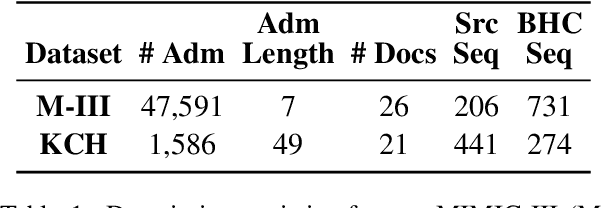
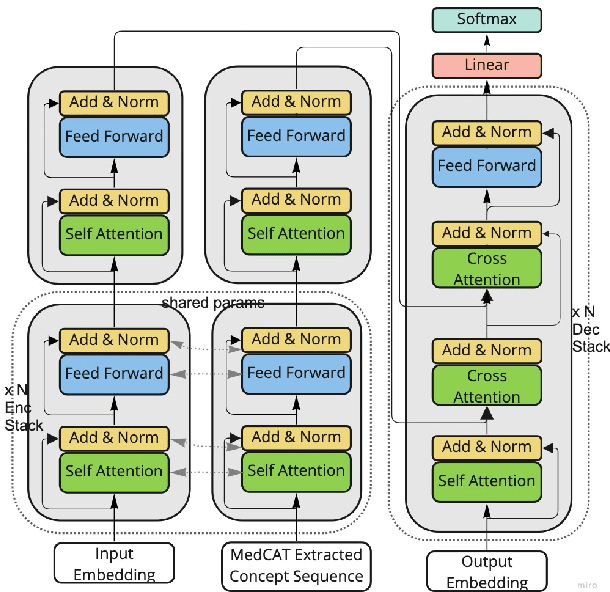
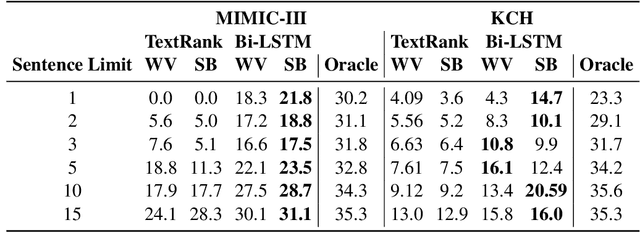
Abstract:Brief Hospital Course (BHC) summaries are succinct summaries of an entire hospital encounter, embedded within discharge summaries, written by senior clinicians responsible for the overall care of a patient. Methods to automatically produce summaries from inpatient documentation would be invaluable in reducing clinician manual burden of summarising documents under high time-pressure to admit and discharge patients. Automatically producing these summaries from the inpatient course, is a complex, multi-document summarisation task, as source notes are written from various perspectives (e.g. nursing, doctor, radiology), during the course of the hospitalisation. We demonstrate a range of methods for BHC summarisation demonstrating the performance of deep learning summarisation models across extractive and abstractive summarisation scenarios. We also test a novel ensemble extractive and abstractive summarisation model that incorporates a medical concept ontology (SNOMED) as a clinical guidance signal and shows superior performance in 2 real-world clinical data sets.
Transformer-based out-of-distribution detection for clinically safe segmentation
May 21, 2022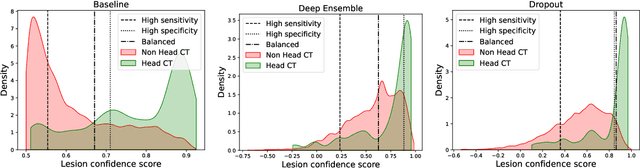
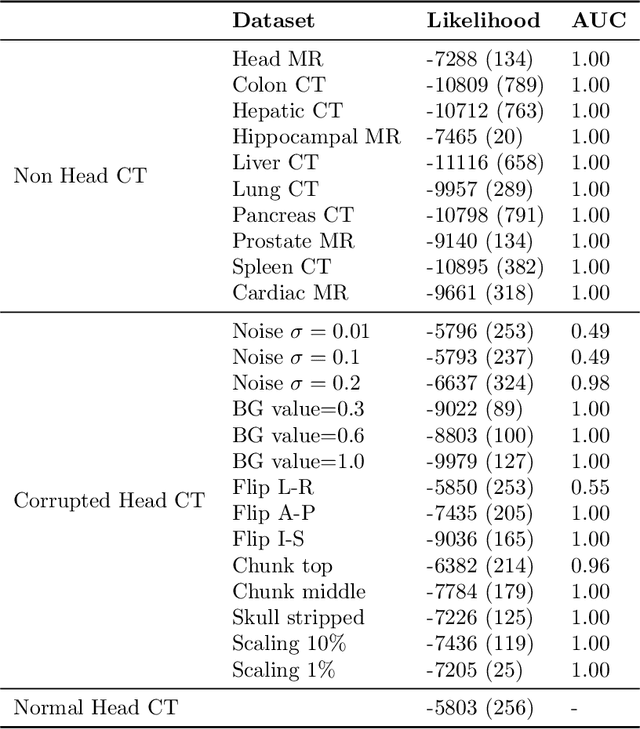
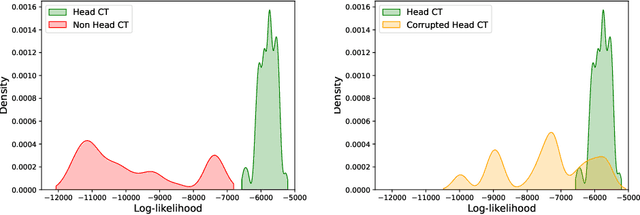
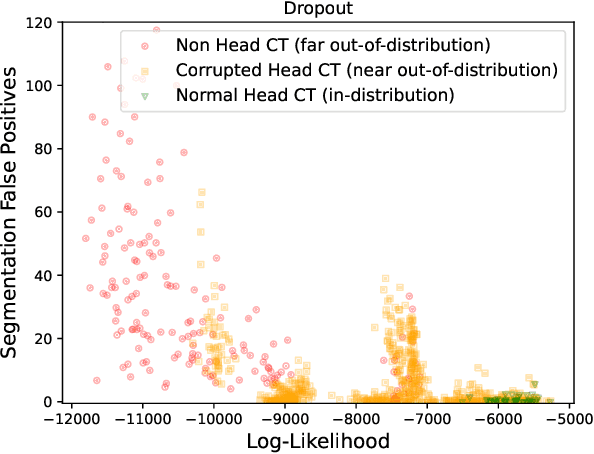
Abstract:In a clinical setting it is essential that deployed image processing systems are robust to the full range of inputs they might encounter and, in particular, do not make confidently wrong predictions. The most popular approach to safe processing is to train networks that can provide a measure of their uncertainty, but these tend to fail for inputs that are far outside the training data distribution. Recently, generative modelling approaches have been proposed as an alternative; these can quantify the likelihood of a data sample explicitly, filtering out any out-of-distribution (OOD) samples before further processing is performed. In this work, we focus on image segmentation and evaluate several approaches to network uncertainty in the far-OOD and near-OOD cases for the task of segmenting haemorrhages in head CTs. We find all of these approaches are unsuitable for safe segmentation as they provide confidently wrong predictions when operating OOD. We propose performing full 3D OOD detection using a VQ-GAN to provide a compressed latent representation of the image and a transformer to estimate the data likelihood. Our approach successfully identifies images in both the far- and near-OOD cases. We find a strong relationship between image likelihood and the quality of a model's segmentation, making this approach viable for filtering images unsuitable for segmentation. To our knowledge, this is the first time transformers have been applied to perform OOD detection on 3D image data.
MedGPT: Medical Concept Prediction from Clinical Narratives
Jul 07, 2021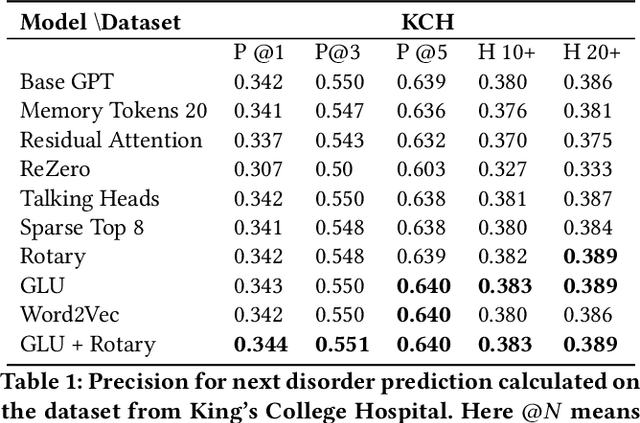
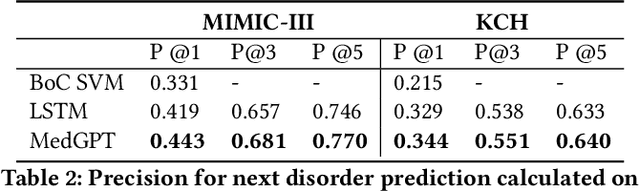
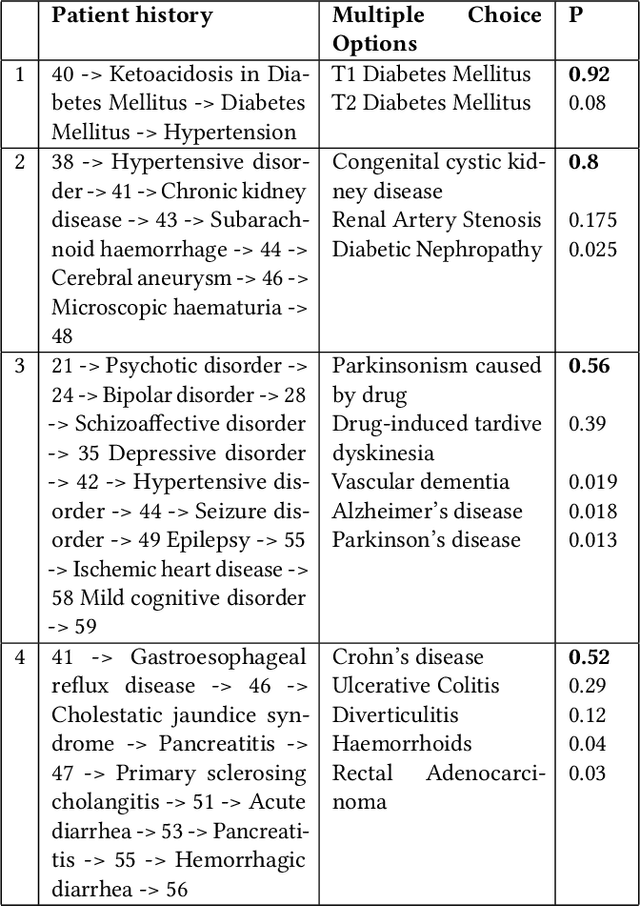
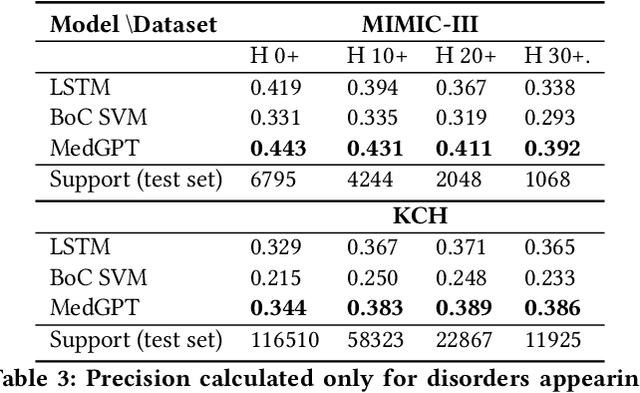
Abstract:The data available in Electronic Health Records (EHRs) provides the opportunity to transform care, and the best way to provide better care for one patient is through learning from the data available on all other patients. Temporal modelling of a patient's medical history, which takes into account the sequence of past events, can be used to predict future events such as a diagnosis of a new disorder or complication of a previous or existing disorder. While most prediction approaches use mostly the structured data in EHRs or a subset of single-domain predictions and outcomes, we present MedGPT a novel transformer-based pipeline that uses Named Entity Recognition and Linking tools (i.e. MedCAT) to structure and organize the free text portion of EHRs and anticipate a range of future medical events (initially disorders). Since a large portion of EHR data is in text form, such an approach benefits from a granular and detailed view of a patient while introducing modest additional noise. MedGPT effectively deals with the noise and the added granularity, and achieves a precision of 0.344, 0.552 and 0.640 (vs LSTM 0.329, 0.538 and 0.633) when predicting the top 1, 3 and 5 candidate future disorders on real world hospital data from King's College Hospital, London, UK (\textasciitilde600k patients). We also show that our model captures medical knowledge by testing it on an experimental medical multiple choice question answering task, and by examining the attentional focus of the model using gradient-based saliency methods.
Estimating Redundancy in Clinical Text
May 25, 2021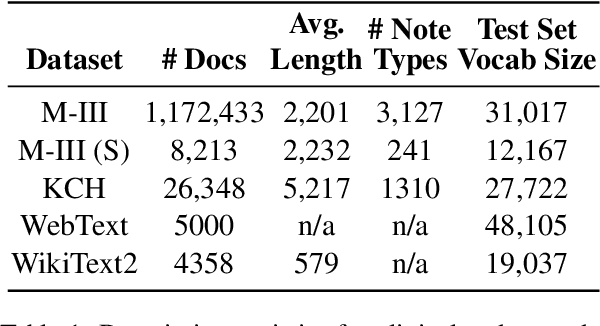
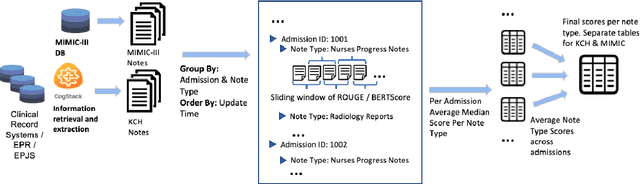
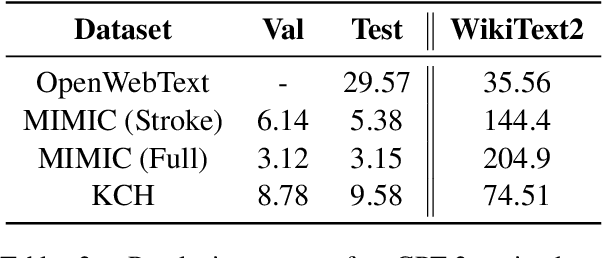
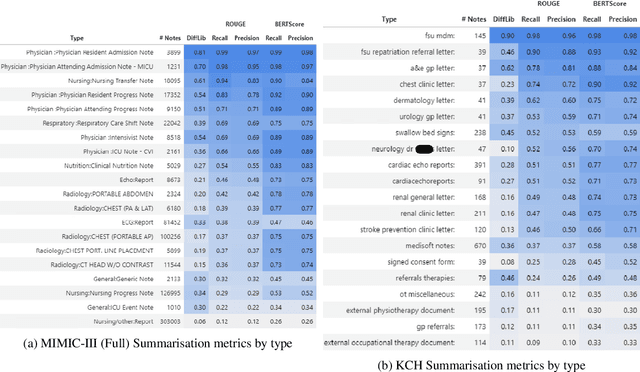
Abstract:The current mode of use of Electronic Health Record (EHR) elicits text redundancy. Clinicians often populate new documents by duplicating existing notes, then updating accordingly. Data duplication can lead to a propagation of errors, inconsistencies and misreporting of care. Therefore, quantifying information redundancy can play an essential role in evaluating innovations that operate on clinical narratives. This work is a quantitative examination of information redundancy in EHR notes. We present and evaluate two strategies to measure redundancy: an information-theoretic approach and a lexicosyntactic and semantic model. We evaluate the measures by training large Transformer-based language models using clinical text from a large openly available US-based ICU dataset and a large multi-site UK based Trust. By comparing the information-theoretic content of the trained models with open-domain language models, the language models trained using clinical text have shown ~1.5x to ~3x less efficient than open-domain corpora. Manual evaluation shows a high correlation with lexicosyntactic and semantic redundancy, with averages ~43 to ~65%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge