Kawsar Noor
Developing and Evaluating an AI-Assisted Prediction Model for Unplanned Intensive Care Admissions following Elective Neurosurgery using Natural Language Processing within an Electronic Healthcare Record System
Mar 13, 2025Abstract:Introduction: Timely care in a specialised neuro-intensive therapy unit (ITU) reduces mortality and hospital stays, with planned admissions being safer than unplanned ones. However, post-operative care decisions remain subjective. This study used artificial intelligence (AI), specifically natural language processing (NLP) to analyse electronic health records (EHRs) and predict ITU admissions for elective surgery patients. Methods: This study analysed the EHRs of elective neurosurgery patients from University College London Hospital (UCLH) using NLP. Patients were categorised into planned high dependency unit (HDU) or ITU admission; unplanned HDU or ITU admission; or ward / overnight recovery (ONR). The Medical Concept Annotation Tool (MedCAT) was used to identify SNOMED-CT concepts within the clinical notes. We then explored the utility of these identified concepts for a range of AI algorithms trained to predict ITU admission. Results: The CogStack-MedCAT NLP model, initially trained on hospital-wide EHRs, underwent two refinements: first with data from patients with Normal Pressure Hydrocephalus (NPH) and then with data from Vestibular Schwannoma (VS) patients, achieving a concept detection F1-score of 0.93. This refined model was then used to extract concepts from EHR notes of 2,268 eligible neurosurgical patients. We integrated the extracted concepts into AI models, including a decision tree model and a neural time-series model. Using the simpler decision tree model, we achieved a recall of 0.87 (CI 0.82 - 0.91) for ITU admissions, reducing the proportion of unplanned ITU cases missed by human experts from 36% to 4%. Conclusion: The NLP model, refined for accuracy, has proven its efficiency in extracting relevant concepts, providing a reliable basis for predictive AI models to use in clinically valid applications.
Validating transformers for redaction of text from electronic health records in real-world healthcare
Oct 05, 2023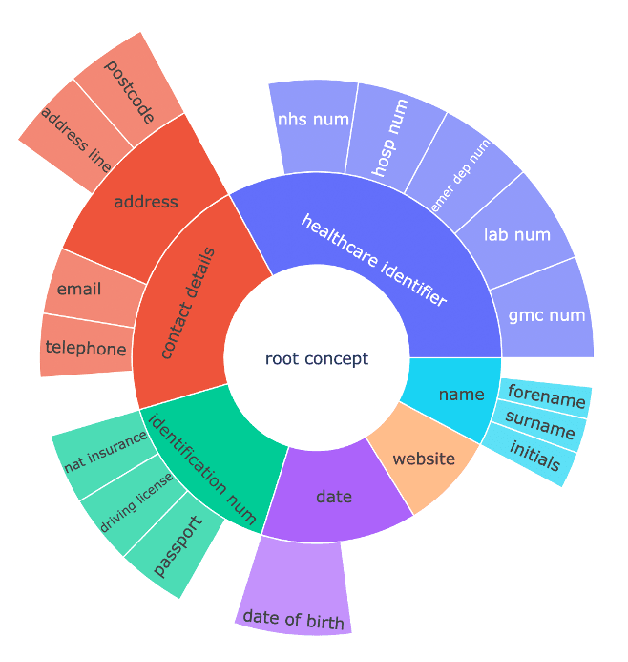
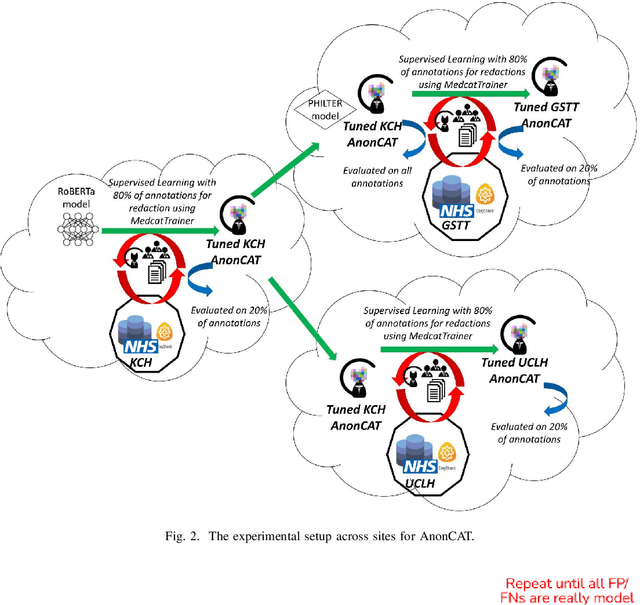
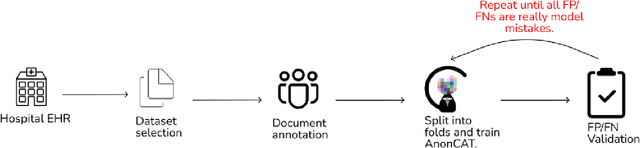
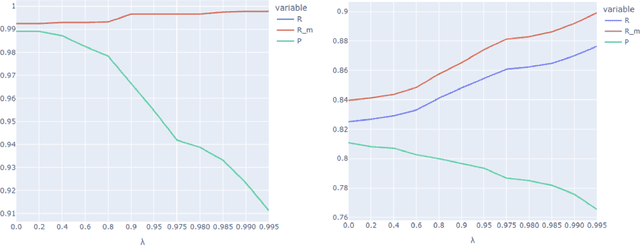
Abstract:Protecting patient privacy in healthcare records is a top priority, and redaction is a commonly used method for obscuring directly identifiable information in text. Rule-based methods have been widely used, but their precision is often low causing over-redaction of text and frequently not being adaptable enough for non-standardised or unconventional structures of personal health information. Deep learning techniques have emerged as a promising solution, but implementing them in real-world environments poses challenges due to the differences in patient record structure and language across different departments, hospitals, and countries. In this study, we present AnonCAT, a transformer-based model and a blueprint on how deidentification models can be deployed in real-world healthcare. AnonCAT was trained through a process involving manually annotated redactions of real-world documents from three UK hospitals with different electronic health record systems and 3116 documents. The model achieved high performance in all three hospitals with a Recall of 0.99, 0.99 and 0.96. Our findings demonstrate the potential of deep learning techniques for improving the efficiency and accuracy of redaction in global healthcare data and highlight the importance of building workflows which not just use these models but are also able to continually fine-tune and audit the performance of these algorithms to ensure continuing effectiveness in real-world settings. This approach provides a blueprint for the real-world use of de-identifying algorithms through fine-tuning and localisation, the code together with tutorials is available on GitHub (https://github.com/CogStack/MedCAT).
Deployment of a Free-Text Analytics Platform at a UK National Health Service Research Hospital: CogStack at University College London Hospitals
Aug 15, 2021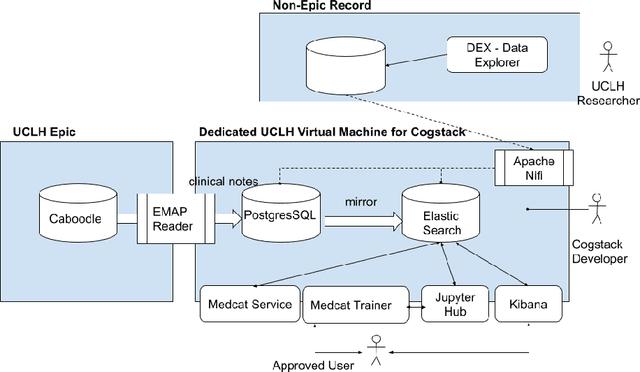
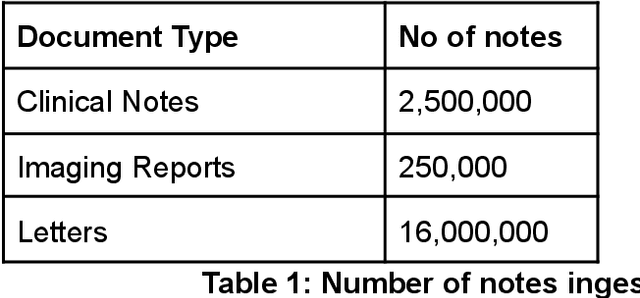

Abstract:As more healthcare organisations transition to using electronic health record (EHR) systems it is important for these organisations to maximise the secondary use of their data to support service improvement and clinical research. These organisations will find it challenging to have systems which can mine information from the unstructured data fields in the record (clinical notes, letters etc) and more practically have such systems interact with all of the hospitals data systems (legacy and current). To tackle this problem at University College London Hospitals, we have deployed an enhanced version of the CogStack platform; an information retrieval platform with natural language processing capabilities which we have configured to process the hospital's existing and legacy records. The platform has improved data ingestion capabilities as well as better tools for natural language processing. To date we have processed over 18 million records and the insights produced from CogStack have informed a number of clinical research use cases at the hospitals.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020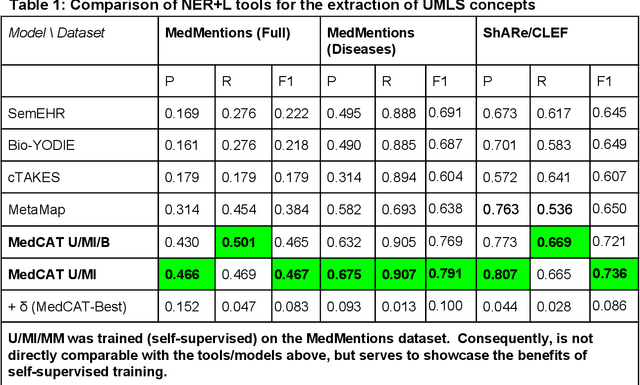
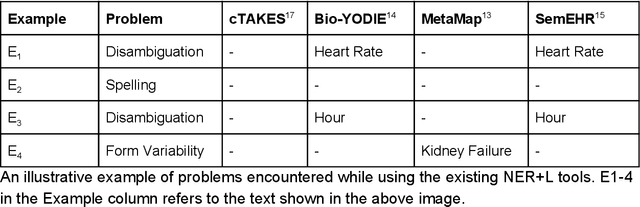
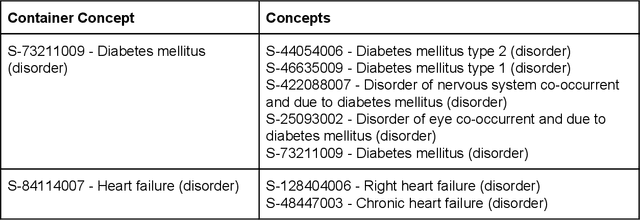
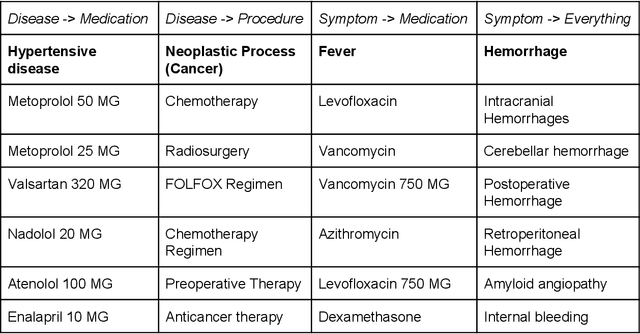
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge