Richard Dobson
Clean & Clear: Feasibility of Safe LLM Clinical Guidance
Mar 26, 2025Abstract:Background: Clinical guidelines are central to safe evidence-based medicine in modern healthcare, providing diagnostic criteria, treatment options and monitoring advice for a wide range of illnesses. LLM-empowered chatbots have shown great promise in Healthcare Q&A tasks, offering the potential to provide quick and accurate responses to medical inquiries. Our main objective was the development and preliminary assessment of an LLM-empowered chatbot software capable of reliably answering clinical guideline questions using University College London Hospital (UCLH) clinical guidelines. Methods: We used the open-weight Llama-3.1-8B LLM to extract relevant information from the UCLH guidelines to answer questions. Our approach highlights the safety and reliability of referencing information over its interpretation and response generation. Seven doctors from the ward assessed the chatbot's performance by comparing its answers to the gold standard. Results: Our chatbot demonstrates promising performance in terms of relevance, with ~73% of its responses rated as very relevant, showcasing a strong understanding of the clinical context. Importantly, our chatbot achieves a recall of 0.98 for extracted guideline lines, substantially minimising the risk of missing critical information. Approximately 78% of responses were rated satisfactory in terms of completeness. A small portion (~14.5%) contained minor unnecessary information, indicating occasional lapses in precision. The chatbot' showed high efficiency, with an average completion time of 10 seconds, compared to 30 seconds for human respondents. Evaluation of clinical reasoning showed that 72% of the chatbot's responses were without flaws. Our chatbot demonstrates significant potential to speed up and improve the process of accessing locally relevant clinical information for healthcare professionals.
RelCAT: Advancing Extraction of Clinical Inter-Entity Relationships from Unstructured Electronic Health Records
Jan 27, 2025Abstract:This study introduces RelCAT (Relation Concept Annotation Toolkit), an interactive tool, library, and workflow designed to classify relations between entities extracted from clinical narratives. Building upon the CogStack MedCAT framework, RelCAT addresses the challenge of capturing complete clinical relations dispersed within text. The toolkit implements state-of-the-art machine learning models such as BERT and Llama along with proven evaluation and training methods. We demonstrate a dataset annotation tool (built within MedCATTrainer), model training, and evaluate our methodology on both openly available gold-standard and real-world UK National Health Service (NHS) hospital clinical datasets. We perform extensive experimentation and a comparative analysis of the various publicly available models with varied approaches selected for model fine-tuning. Finally, we achieve macro F1-scores of 0.977 on the gold-standard n2c2, surpassing the previous state-of-the-art performance, and achieve performance of >=0.93 F1 on our NHS gathered datasets.
Improving Extraction of Clinical Event Contextual Properties from Electronic Health Records: A Comparative Study
Aug 30, 2024
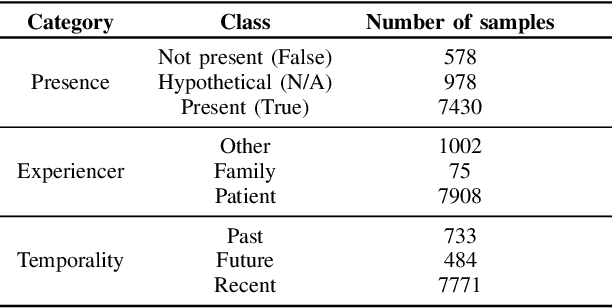
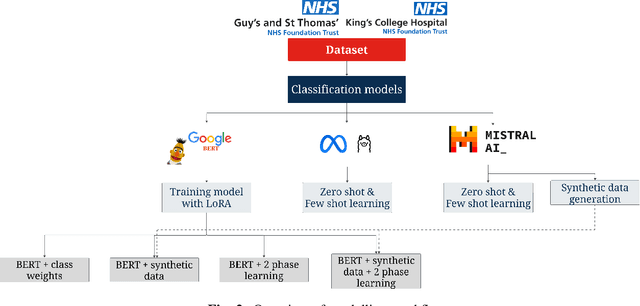
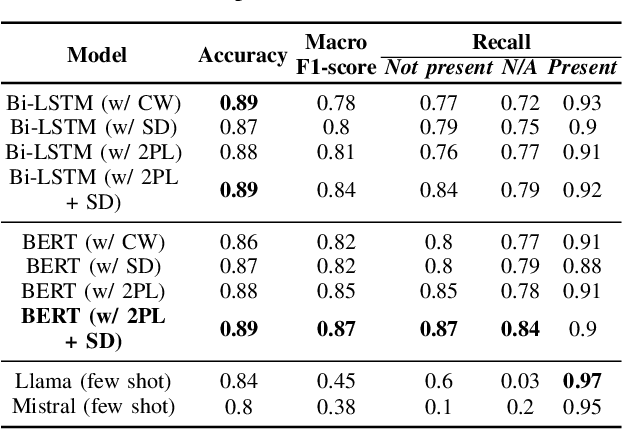
Abstract:Electronic Health Records are large repositories of valuable clinical data, with a significant portion stored in unstructured text format. This textual data includes clinical events (e.g., disorders, symptoms, findings, medications and procedures) in context that if extracted accurately at scale can unlock valuable downstream applications such as disease prediction. Using an existing Named Entity Recognition and Linking methodology, MedCAT, these identified concepts need to be further classified (contextualised) for their relevance to the patient, and their temporal and negated status for example, to be useful downstream. This study performs a comparative analysis of various natural language models for medical text classification. Extensive experimentation reveals the effectiveness of transformer-based language models, particularly BERT. When combined with class imbalance mitigation techniques, BERT outperforms Bi-LSTM models by up to 28% and the baseline BERT model by up to 16% for recall of the minority classes. The method has been implemented as part of CogStack/MedCAT framework and made available to the community for further research.
How Deep is your Guess? A Fresh Perspective on Deep Learning for Medical Time-Series Imputation
Jul 11, 2024Abstract:We introduce a novel classification framework for time-series imputation using deep learning, with a particular focus on clinical data. By identifying conceptual gaps in the literature and existing reviews, we devise a taxonomy grounded on the inductive bias of neural imputation frameworks, resulting in a classification of existing deep imputation strategies based on their suitability for specific imputation scenarios and data-specific properties. Our review further examines the existing methodologies employed to benchmark deep imputation models, evaluating their effectiveness in capturing the missingness scenarios found in clinical data and emphasising the importance of reconciling mathematical abstraction with clinical insights. Our classification aims to serve as a guide for researchers to facilitate the selection of appropriate deep learning imputation techniques tailored to their specific clinical data. Our novel perspective also highlights the significance of bridging the gap between computational methodologies and medical insights to achieve clinically sound imputation models.
Knowledge Enhanced Conditional Imputation for Healthcare Time-series
Jan 04, 2024Abstract:This study presents a novel approach to addressing the challenge of missing data in multivariate time series, with a particular focus on the complexities of healthcare data. Our Conditional Self-Attention Imputation (CSAI) model, grounded in a transformer-based framework, introduces a conditional hidden state initialization tailored to the intricacies of medical time series data. This methodology diverges from traditional imputation techniques by specifically targeting the imbalance in missing data distribution, a crucial aspect often overlooked in healthcare datasets. By integrating advanced knowledge embedding and a non-uniform masking strategy, CSAI adeptly adjusts to the distinct patterns of missing data in Electronic Health Records (EHRs).
Uncertainty-Aware Deep Attention Recurrent Neural Network for Heterogeneous Time Series Imputation
Jan 04, 2024Abstract:Missingness is ubiquitous in multivariate time series and poses an obstacle to reliable downstream analysis. Although recurrent network imputation achieved the SOTA, existing models do not scale to deep architectures that can potentially alleviate issues arising in complex data. Moreover, imputation carries the risk of biased estimations of the ground truth. Yet, confidence in the imputed values is always unmeasured or computed post hoc from model output. We propose DEep Attention Recurrent Imputation (DEARI), which jointly estimates missing values and their associated uncertainty in heterogeneous multivariate time series. By jointly representing feature-wise correlations and temporal dynamics, we adopt a self attention mechanism, along with an effective residual component, to achieve a deep recurrent neural network with good imputation performance and stable convergence. We also leverage self-supervised metric learning to boost performance by optimizing sample similarity. Finally, we transform DEARI into a Bayesian neural network through a novel Bayesian marginalization strategy to produce stochastic DEARI, which outperforms its deterministic equivalent. Experiments show that DEARI surpasses the SOTA in diverse imputation tasks using real-world datasets, namely air quality control, healthcare and traffic.
Validating transformers for redaction of text from electronic health records in real-world healthcare
Oct 05, 2023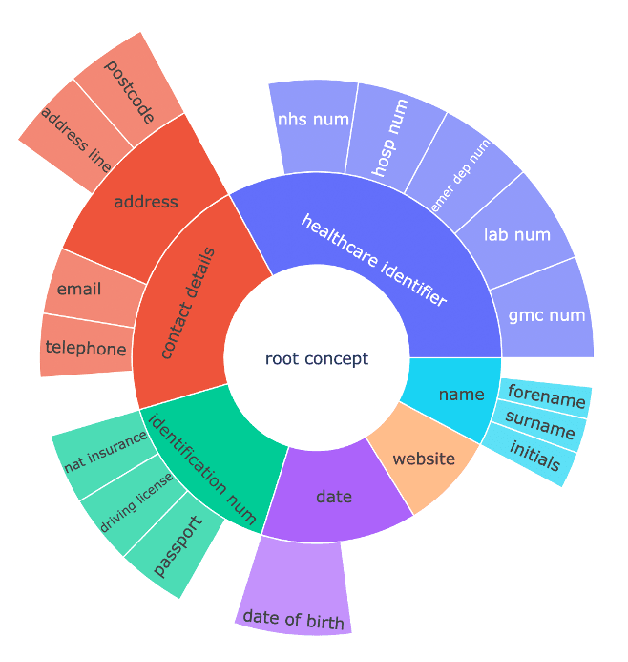
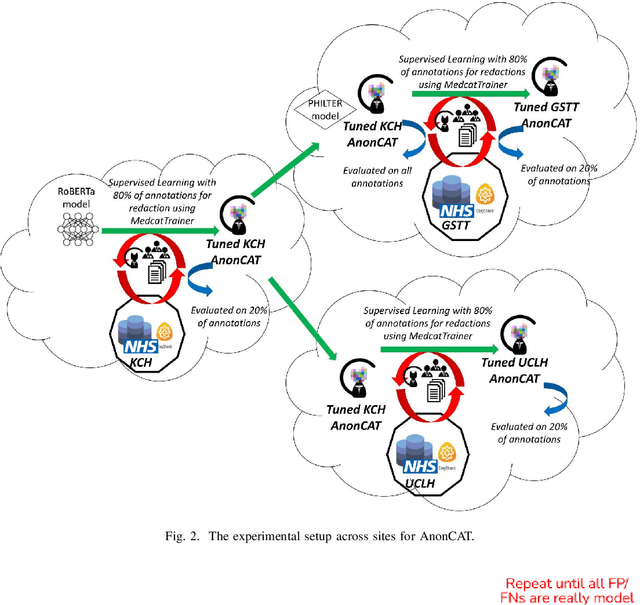
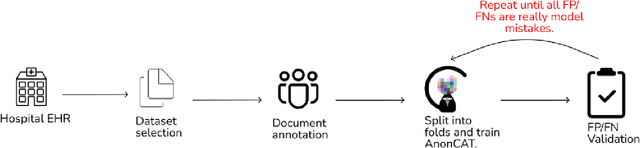
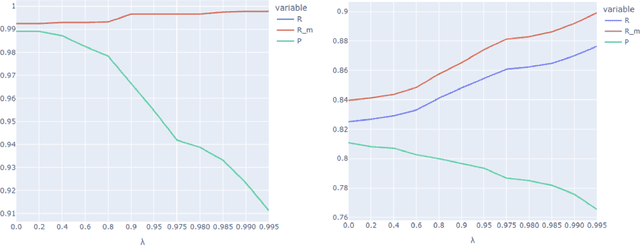
Abstract:Protecting patient privacy in healthcare records is a top priority, and redaction is a commonly used method for obscuring directly identifiable information in text. Rule-based methods have been widely used, but their precision is often low causing over-redaction of text and frequently not being adaptable enough for non-standardised or unconventional structures of personal health information. Deep learning techniques have emerged as a promising solution, but implementing them in real-world environments poses challenges due to the differences in patient record structure and language across different departments, hospitals, and countries. In this study, we present AnonCAT, a transformer-based model and a blueprint on how deidentification models can be deployed in real-world healthcare. AnonCAT was trained through a process involving manually annotated redactions of real-world documents from three UK hospitals with different electronic health record systems and 3116 documents. The model achieved high performance in all three hospitals with a Recall of 0.99, 0.99 and 0.96. Our findings demonstrate the potential of deep learning techniques for improving the efficiency and accuracy of redaction in global healthcare data and highlight the importance of building workflows which not just use these models but are also able to continually fine-tune and audit the performance of these algorithms to ensure continuing effectiveness in real-world settings. This approach provides a blueprint for the real-world use of de-identifying algorithms through fine-tuning and localisation, the code together with tutorials is available on GitHub (https://github.com/CogStack/MedCAT).
Towards robust paralinguistic assessment for real-world mobile health (mHealth) monitoring: an initial study of reverberation effects on speech
May 21, 2023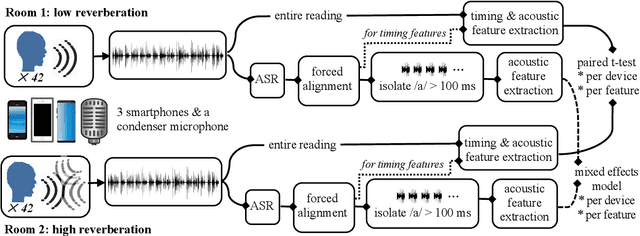

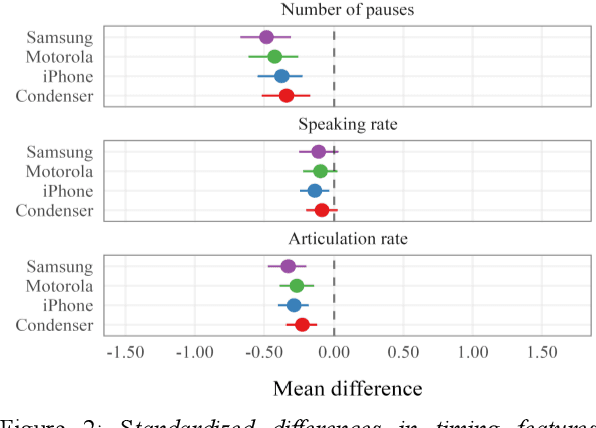
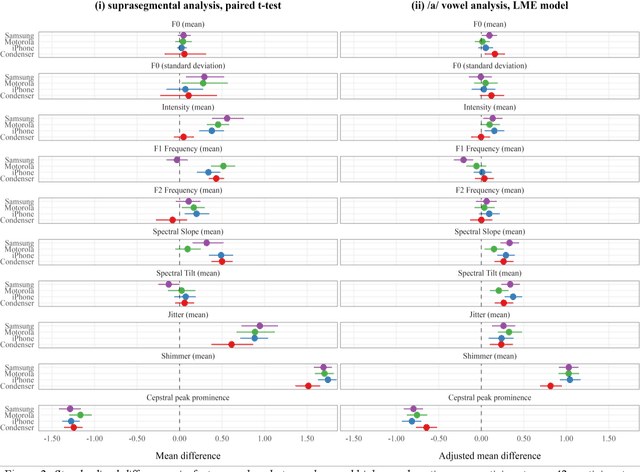
Abstract:Speech is promising as an objective, convenient tool to monitor health remotely over time using mobile devices. Numerous paralinguistic features have been demonstrated to contain salient information related to an individual's health. However, mobile device specification and acoustic environments vary widely, risking the reliability of the extracted features. In an initial step towards quantifying these effects, we report the variability of 13 exemplar paralinguistic features commonly reported in the speech-health literature and extracted from the speech of 42 healthy volunteers recorded consecutively in rooms with low and high reverberation with one budget and two higher-end smartphones and a condenser microphone. Our results show reverberation has a clear effect on several features, in particular voice quality markers. They point to new research directions investigating how best to record and process in-the-wild speech for reliable longitudinal health state assessment.
Summarisation of Electronic Health Records with Clinical Concept Guidance
Nov 14, 2022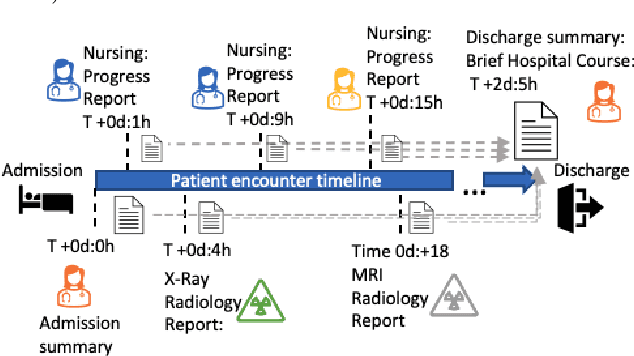
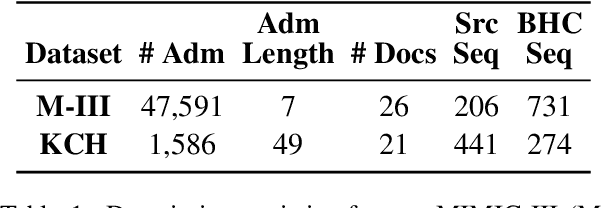
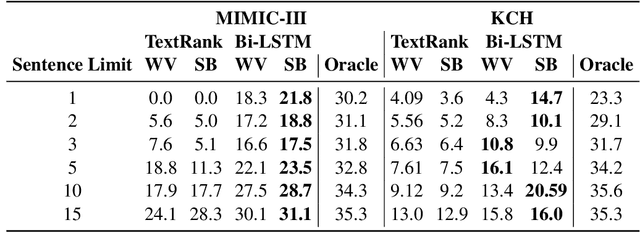
Abstract:Brief Hospital Course (BHC) summaries are succinct summaries of an entire hospital encounter, embedded within discharge summaries, written by senior clinicians responsible for the overall care of a patient. Methods to automatically produce summaries from inpatient documentation would be invaluable in reducing clinician manual burden of summarising documents under high time-pressure to admit and discharge patients. Automatically producing these summaries from the inpatient course, is a complex, multi-document summarisation task, as source notes are written from various perspectives (e.g. nursing, doctor, radiology), during the course of the hospitalisation. We demonstrate a range of methods for BHC summarisation demonstrating the performance of deep learning summarisation models across extractive and abstractive summarisation scenarios. We also test a novel ensemble extractive and abstractive summarisation model that incorporates a medical concept ontology (SNOMED) as a clinical guidance signal and shows superior performance in 2 real-world clinical data sets.
MedGPT: Medical Concept Prediction from Clinical Narratives
Jul 07, 2021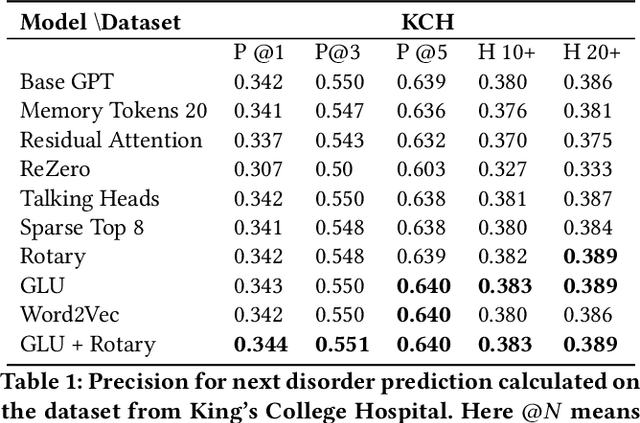
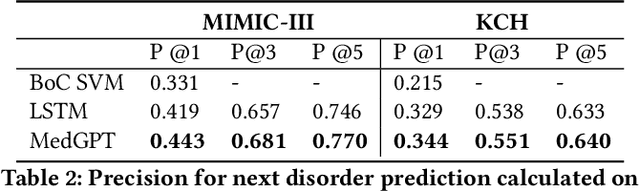
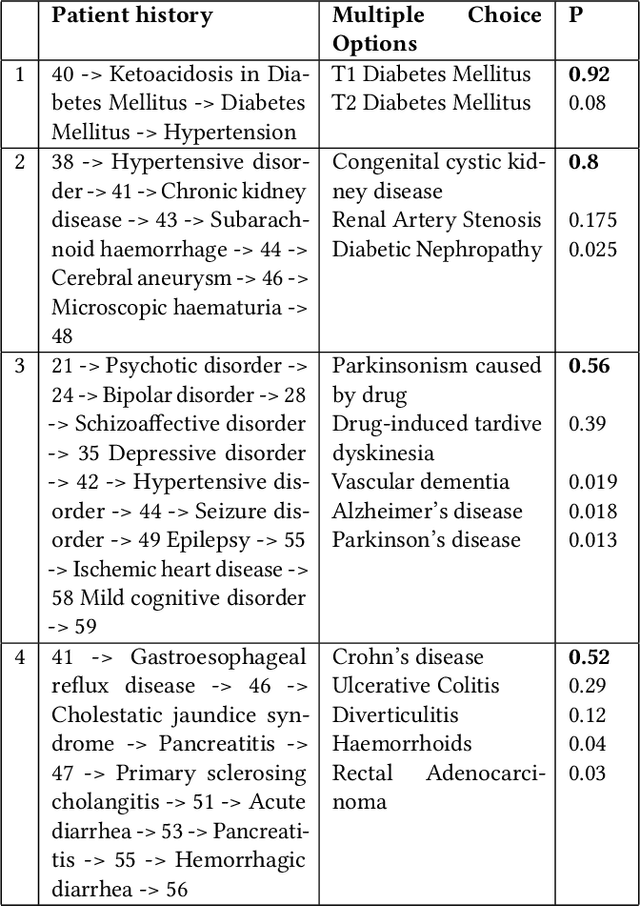
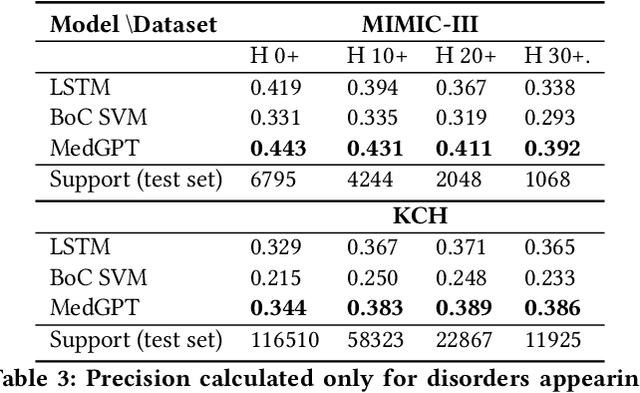
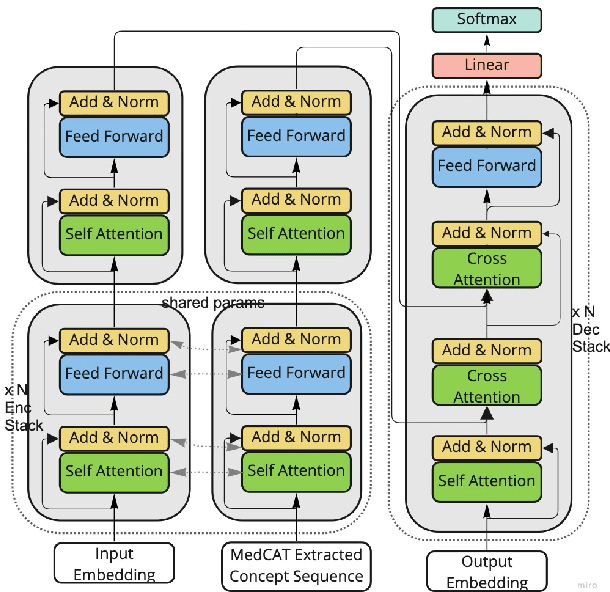
Abstract:The data available in Electronic Health Records (EHRs) provides the opportunity to transform care, and the best way to provide better care for one patient is through learning from the data available on all other patients. Temporal modelling of a patient's medical history, which takes into account the sequence of past events, can be used to predict future events such as a diagnosis of a new disorder or complication of a previous or existing disorder. While most prediction approaches use mostly the structured data in EHRs or a subset of single-domain predictions and outcomes, we present MedGPT a novel transformer-based pipeline that uses Named Entity Recognition and Linking tools (i.e. MedCAT) to structure and organize the free text portion of EHRs and anticipate a range of future medical events (initially disorders). Since a large portion of EHR data is in text form, such an approach benefits from a granular and detailed view of a patient while introducing modest additional noise. MedGPT effectively deals with the noise and the added granularity, and achieves a precision of 0.344, 0.552 and 0.640 (vs LSTM 0.329, 0.538 and 0.633) when predicting the top 1, 3 and 5 candidate future disorders on real world hospital data from King's College Hospital, London, UK (\textasciitilde600k patients). We also show that our model captures medical knowledge by testing it on an experimental medical multiple choice question answering task, and by examining the attentional focus of the model using gradient-based saliency methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge