Paulina Bondaronek
Clean & Clear: Feasibility of Safe LLM Clinical Guidance
Mar 26, 2025Abstract:Background: Clinical guidelines are central to safe evidence-based medicine in modern healthcare, providing diagnostic criteria, treatment options and monitoring advice for a wide range of illnesses. LLM-empowered chatbots have shown great promise in Healthcare Q&A tasks, offering the potential to provide quick and accurate responses to medical inquiries. Our main objective was the development and preliminary assessment of an LLM-empowered chatbot software capable of reliably answering clinical guideline questions using University College London Hospital (UCLH) clinical guidelines. Methods: We used the open-weight Llama-3.1-8B LLM to extract relevant information from the UCLH guidelines to answer questions. Our approach highlights the safety and reliability of referencing information over its interpretation and response generation. Seven doctors from the ward assessed the chatbot's performance by comparing its answers to the gold standard. Results: Our chatbot demonstrates promising performance in terms of relevance, with ~73% of its responses rated as very relevant, showcasing a strong understanding of the clinical context. Importantly, our chatbot achieves a recall of 0.98 for extracted guideline lines, substantially minimising the risk of missing critical information. Approximately 78% of responses were rated satisfactory in terms of completeness. A small portion (~14.5%) contained minor unnecessary information, indicating occasional lapses in precision. The chatbot' showed high efficiency, with an average completion time of 10 seconds, compared to 30 seconds for human respondents. Evaluation of clinical reasoning showed that 72% of the chatbot's responses were without flaws. Our chatbot demonstrates significant potential to speed up and improve the process of accessing locally relevant clinical information for healthcare professionals.
LLM Assistance for Pediatric Depression
Jan 29, 2025



Abstract:Traditional depression screening methods, such as the PHQ-9, are particularly challenging for children in pediatric primary care due to practical limitations. AI has the potential to help, but the scarcity of annotated datasets in mental health, combined with the computational costs of training, highlights the need for efficient, zero-shot approaches. In this work, we investigate the feasibility of state-of-the-art LLMs for depressive symptom extraction in pediatric settings (ages 6-24). This approach aims to complement traditional screening and minimize diagnostic errors. Our findings show that all LLMs are 60% more efficient than word match, with Flan leading in precision (average F1: 0.65, precision: 0.78), excelling in the extraction of more rare symptoms like "sleep problems" (F1: 0.92) and "self-loathing" (F1: 0.8). Phi strikes a balance between precision (0.44) and recall (0.60), performing well in categories like "Feeling depressed" (0.69) and "Weight change" (0.78). Llama 3, with the highest recall (0.90), overgeneralizes symptoms, making it less suitable for this type of analysis. Challenges include the complexity of clinical notes and overgeneralization from PHQ-9 scores. The main challenges faced by LLMs include navigating the complex structure of clinical notes with content from different times in the patient trajectory, as well as misinterpreting elevated PHQ-9 scores. We finally demonstrate the utility of symptom annotations provided by Flan as features in an ML algorithm, which differentiates depression cases from controls with high precision of 0.78, showing a major performance boost compared to a baseline that does not use these features.
A Data-Centric Approach to Detecting and Mitigating Demographic Bias in Pediatric Mental Health Text: A Case Study in Anxiety Detection
Dec 30, 2024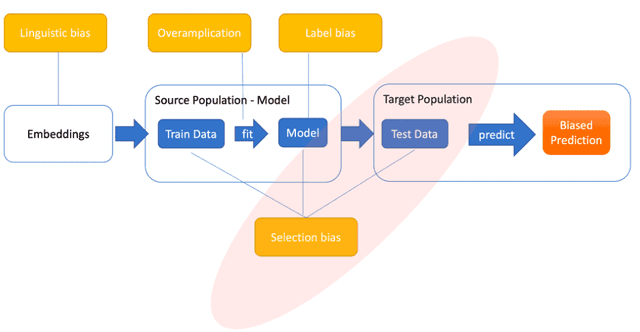
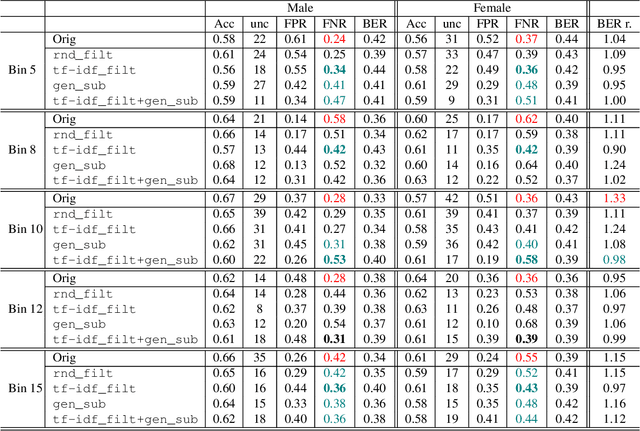
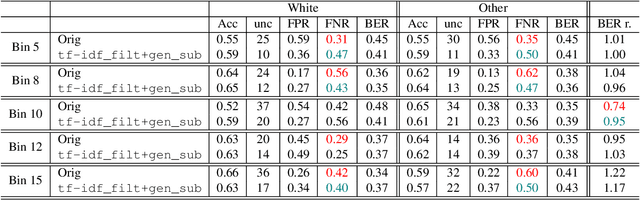
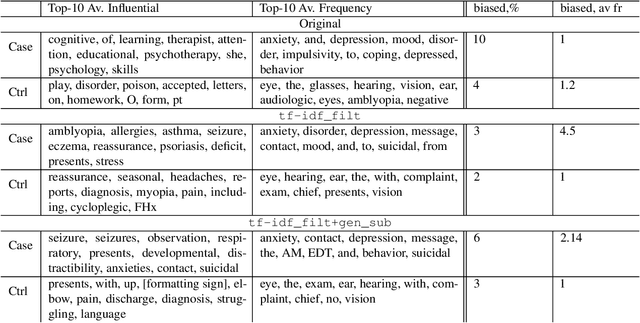
Abstract:Introduction: Healthcare AI models often inherit biases from their training data. While efforts have primarily targeted bias in structured data, mental health heavily depends on unstructured data. This study aims to detect and mitigate linguistic differences related to non-biological differences in the training data of AI models designed to assist in pediatric mental health screening. Our objectives are: (1) to assess the presence of bias by evaluating outcome parity across sex subgroups, (2) to identify bias sources through textual distribution analysis, and (3) to develop a de-biasing method for mental health text data. Methods: We examined classification parity across demographic groups and assessed how gendered language influences model predictions. A data-centric de-biasing method was applied, focusing on neutralizing biased terms while retaining salient clinical information. This methodology was tested on a model for automatic anxiety detection in pediatric patients. Results: Our findings revealed a systematic under-diagnosis of female adolescent patients, with a 4% lower accuracy and a 9% higher False Negative Rate (FNR) compared to male patients, likely due to disparities in information density and linguistic differences in patient notes. Notes for male patients were on average 500 words longer, and linguistic similarity metrics indicated distinct word distributions between genders. Implementing our de-biasing approach reduced diagnostic bias by up to 27%, demonstrating its effectiveness in enhancing equity across demographic groups. Discussion: We developed a data-centric de-biasing framework to address gender-based content disparities within clinical text. By neutralizing biased language and enhancing focus on clinically essential information, our approach demonstrates an effective strategy for mitigating bias in AI healthcare models trained on text.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge