Julia Ive
Context-Emotion Aware Therapeutic Dialogue Generation: A Multi-component Reinforcement Learning Approach to Language Models for Mental Health Support
Nov 14, 2025Abstract:Mental health illness represents a substantial global socioeconomic burden, with COVID-19 further exacerbating accessibility challenges and driving increased demand for telehealth mental health support. While large language models (LLMs) offer promising solutions through 24/7 availability and non-judgmental interactions, pre-trained models often lack the contextual and emotional awareness necessary for appropriate therapeutic responses. This paper investigated the application of supervised fine-tuning (SFT) and reinforcement learning (RL) techniques to enhance GPT-2's capacity for therapeutic dialogue generation. The methodology restructured input formats to enable simultaneous processing of contextual information and emotional states alongside user input, employing a multi-component reward function that aligned model outputs with professional therapist responses and annotated emotions. Results demonstrated improvements through reinforcement learning over baseline GPT-2 across multiple evaluation metrics: BLEU (0.0111), ROUGE-1 (0.1397), ROUGE-2 (0.0213), ROUGE-L (0.1317), and METEOR (0.0581). LLM evaluation confirmed high contextual relevance and professionalism, while reinforcement learning achieved 99.34% emotion accuracy compared to 66.96% for baseline GPT-2. These findings demonstrate reinforcement learning's effectiveness in developing therapeutic dialogue systems that can serve as valuable assistive tools for therapists while maintaining essential human clinical oversight.
Hesitation is defeat? Connecting Linguistic and Predictive Uncertainty
May 06, 2025Abstract:Automating chest radiograph interpretation using Deep Learning (DL) models has the potential to significantly improve clinical workflows, decision-making, and large-scale health screening. However, in medical settings, merely optimising predictive performance is insufficient, as the quantification of uncertainty is equally crucial. This paper investigates the relationship between predictive uncertainty, derived from Bayesian Deep Learning approximations, and human/linguistic uncertainty, as estimated from free-text radiology reports labelled by rule-based labellers. Utilising BERT as the model of choice, this study evaluates different binarisation methods for uncertainty labels and explores the efficacy of Monte Carlo Dropout and Deep Ensembles in estimating predictive uncertainty. The results demonstrate good model performance, but also a modest correlation between predictive and linguistic uncertainty, highlighting the challenges in aligning machine uncertainty with human interpretation nuances. Our findings suggest that while Bayesian approximations provide valuable uncertainty estimates, further refinement is necessary to fully capture and utilise the subtleties of human uncertainty in clinical applications.
Clean & Clear: Feasibility of Safe LLM Clinical Guidance
Mar 26, 2025Abstract:Background: Clinical guidelines are central to safe evidence-based medicine in modern healthcare, providing diagnostic criteria, treatment options and monitoring advice for a wide range of illnesses. LLM-empowered chatbots have shown great promise in Healthcare Q&A tasks, offering the potential to provide quick and accurate responses to medical inquiries. Our main objective was the development and preliminary assessment of an LLM-empowered chatbot software capable of reliably answering clinical guideline questions using University College London Hospital (UCLH) clinical guidelines. Methods: We used the open-weight Llama-3.1-8B LLM to extract relevant information from the UCLH guidelines to answer questions. Our approach highlights the safety and reliability of referencing information over its interpretation and response generation. Seven doctors from the ward assessed the chatbot's performance by comparing its answers to the gold standard. Results: Our chatbot demonstrates promising performance in terms of relevance, with ~73% of its responses rated as very relevant, showcasing a strong understanding of the clinical context. Importantly, our chatbot achieves a recall of 0.98 for extracted guideline lines, substantially minimising the risk of missing critical information. Approximately 78% of responses were rated satisfactory in terms of completeness. A small portion (~14.5%) contained minor unnecessary information, indicating occasional lapses in precision. The chatbot' showed high efficiency, with an average completion time of 10 seconds, compared to 30 seconds for human respondents. Evaluation of clinical reasoning showed that 72% of the chatbot's responses were without flaws. Our chatbot demonstrates significant potential to speed up and improve the process of accessing locally relevant clinical information for healthcare professionals.
Developing and Evaluating an AI-Assisted Prediction Model for Unplanned Intensive Care Admissions following Elective Neurosurgery using Natural Language Processing within an Electronic Healthcare Record System
Mar 13, 2025Abstract:Introduction: Timely care in a specialised neuro-intensive therapy unit (ITU) reduces mortality and hospital stays, with planned admissions being safer than unplanned ones. However, post-operative care decisions remain subjective. This study used artificial intelligence (AI), specifically natural language processing (NLP) to analyse electronic health records (EHRs) and predict ITU admissions for elective surgery patients. Methods: This study analysed the EHRs of elective neurosurgery patients from University College London Hospital (UCLH) using NLP. Patients were categorised into planned high dependency unit (HDU) or ITU admission; unplanned HDU or ITU admission; or ward / overnight recovery (ONR). The Medical Concept Annotation Tool (MedCAT) was used to identify SNOMED-CT concepts within the clinical notes. We then explored the utility of these identified concepts for a range of AI algorithms trained to predict ITU admission. Results: The CogStack-MedCAT NLP model, initially trained on hospital-wide EHRs, underwent two refinements: first with data from patients with Normal Pressure Hydrocephalus (NPH) and then with data from Vestibular Schwannoma (VS) patients, achieving a concept detection F1-score of 0.93. This refined model was then used to extract concepts from EHR notes of 2,268 eligible neurosurgical patients. We integrated the extracted concepts into AI models, including a decision tree model and a neural time-series model. Using the simpler decision tree model, we achieved a recall of 0.87 (CI 0.82 - 0.91) for ITU admissions, reducing the proportion of unplanned ITU cases missed by human experts from 36% to 4%. Conclusion: The NLP model, refined for accuracy, has proven its efficiency in extracting relevant concepts, providing a reliable basis for predictive AI models to use in clinically valid applications.
LLM Assistance for Pediatric Depression
Jan 29, 2025



Abstract:Traditional depression screening methods, such as the PHQ-9, are particularly challenging for children in pediatric primary care due to practical limitations. AI has the potential to help, but the scarcity of annotated datasets in mental health, combined with the computational costs of training, highlights the need for efficient, zero-shot approaches. In this work, we investigate the feasibility of state-of-the-art LLMs for depressive symptom extraction in pediatric settings (ages 6-24). This approach aims to complement traditional screening and minimize diagnostic errors. Our findings show that all LLMs are 60% more efficient than word match, with Flan leading in precision (average F1: 0.65, precision: 0.78), excelling in the extraction of more rare symptoms like "sleep problems" (F1: 0.92) and "self-loathing" (F1: 0.8). Phi strikes a balance between precision (0.44) and recall (0.60), performing well in categories like "Feeling depressed" (0.69) and "Weight change" (0.78). Llama 3, with the highest recall (0.90), overgeneralizes symptoms, making it less suitable for this type of analysis. Challenges include the complexity of clinical notes and overgeneralization from PHQ-9 scores. The main challenges faced by LLMs include navigating the complex structure of clinical notes with content from different times in the patient trajectory, as well as misinterpreting elevated PHQ-9 scores. We finally demonstrate the utility of symptom annotations provided by Flan as features in an ML algorithm, which differentiates depression cases from controls with high precision of 0.78, showing a major performance boost compared to a baseline that does not use these features.
A Data-Centric Approach to Detecting and Mitigating Demographic Bias in Pediatric Mental Health Text: A Case Study in Anxiety Detection
Dec 30, 2024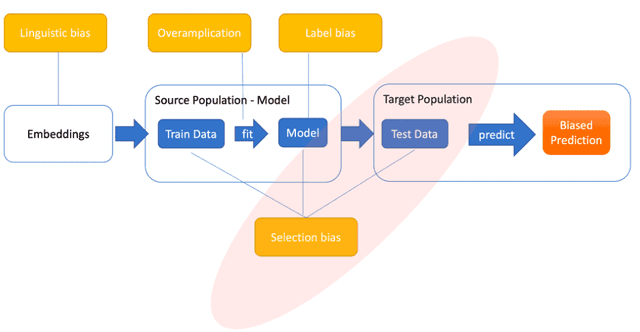
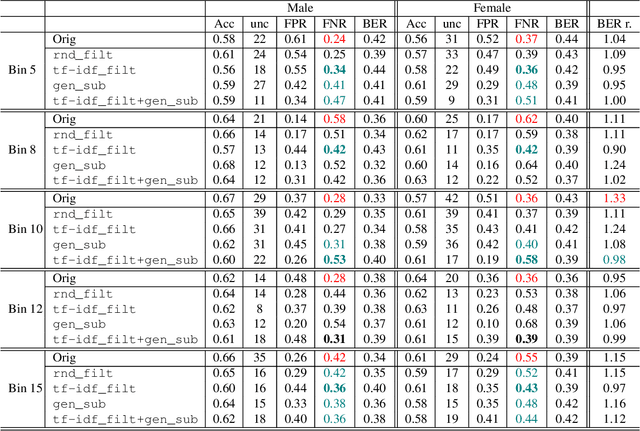
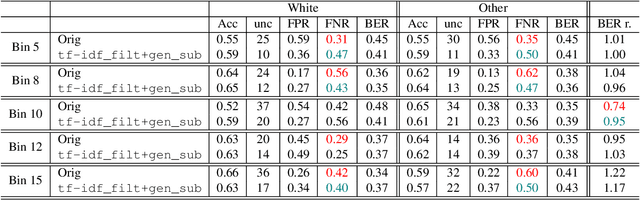
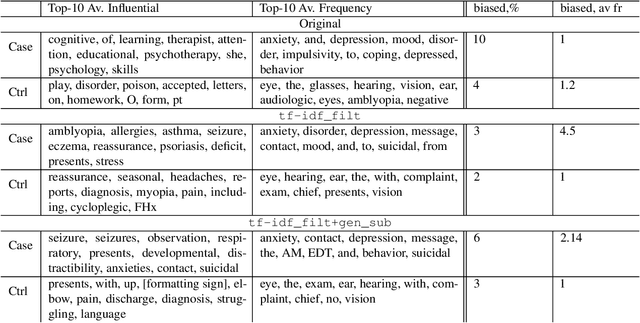
Abstract:Introduction: Healthcare AI models often inherit biases from their training data. While efforts have primarily targeted bias in structured data, mental health heavily depends on unstructured data. This study aims to detect and mitigate linguistic differences related to non-biological differences in the training data of AI models designed to assist in pediatric mental health screening. Our objectives are: (1) to assess the presence of bias by evaluating outcome parity across sex subgroups, (2) to identify bias sources through textual distribution analysis, and (3) to develop a de-biasing method for mental health text data. Methods: We examined classification parity across demographic groups and assessed how gendered language influences model predictions. A data-centric de-biasing method was applied, focusing on neutralizing biased terms while retaining salient clinical information. This methodology was tested on a model for automatic anxiety detection in pediatric patients. Results: Our findings revealed a systematic under-diagnosis of female adolescent patients, with a 4% lower accuracy and a 9% higher False Negative Rate (FNR) compared to male patients, likely due to disparities in information density and linguistic differences in patient notes. Notes for male patients were on average 500 words longer, and linguistic similarity metrics indicated distinct word distributions between genders. Implementing our de-biasing approach reduced diagnostic bias by up to 27%, demonstrating its effectiveness in enhancing equity across demographic groups. Discussion: We developed a data-centric de-biasing framework to address gender-based content disparities within clinical text. By neutralizing biased language and enhancing focus on clinically essential information, our approach demonstrates an effective strategy for mitigating bias in AI healthcare models trained on text.
Safe Training with Sensitive In-domain Data: Leveraging Data Fragmentation To Mitigate Linkage Attacks
Apr 30, 2024Abstract:Current text generation models are trained using real data which can potentially contain sensitive information, such as confidential patient information and the like. Under certain conditions output of the training data which they have memorised can be triggered, exposing sensitive data. To mitigate against this risk we propose a safer alternative which sees fragmented data in the form of domain-specific short phrases randomly grouped together shared instead of full texts. Thus, text fragments that could re-identify an individual cannot be reproduced by the model in one sequence, giving significant protection against linkage attacks. We fine-tune several state-of-the-art LLMs using meaningful syntactic chunks to explore their utility. In particular, we fine-tune BERT-based models to predict two cardiovascular diagnoses. Our results demonstrate the capacity of LLMs to benefit from the pre-trained knowledge and deliver classification results when fine-tuned with fragmented data comparable to fine-tuning with full training data.
Clinically meaningful timeline summarisation in social media for mental health monitoring
Jan 29, 2024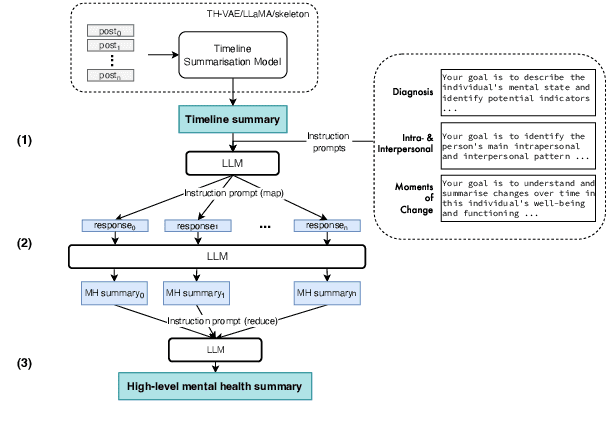


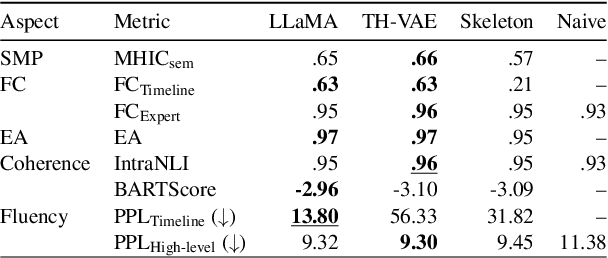
Abstract:We introduce the new task of clinically meaningful summarisation of social media user timelines, appropriate for mental health monitoring. We develop a novel approach for unsupervised abstractive summarisation that produces a two-layer summary consisting of both high-level information, covering aspects useful to clinical experts, as well as accompanying time sensitive evidence from a user's social media timeline. A key methodological novelty comes from the timeline summarisation component based on a version of hierarchical variational autoencoder (VAE) adapted to represent long texts and guided by LLM-annotated key phrases. The resulting timeline summary is input into a LLM (LLaMA-2) to produce the final summary containing both the high level information, obtained through instruction prompting, as well as corresponding evidence from the user's timeline. We assess the summaries generated by our novel architecture via automatic evaluation against expert written summaries and via human evaluation with clinical experts, showing that timeline summarisation by TH-VAE results in logically coherent summaries rich in clinical utility and superior to LLM-only approaches in capturing changes over time.
Source Code is a Graph, Not a Sequence: A Cross-Lingual Perspective on Code Clone Detection
Dec 27, 2023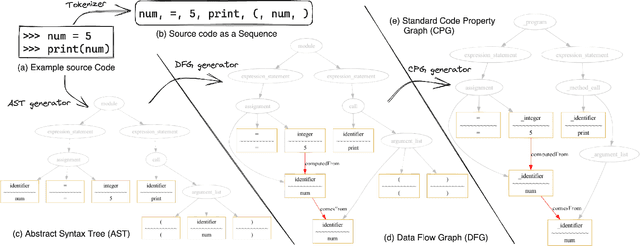
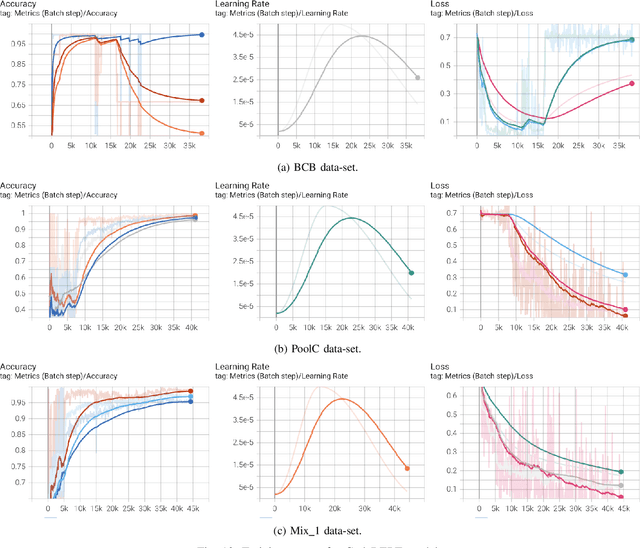
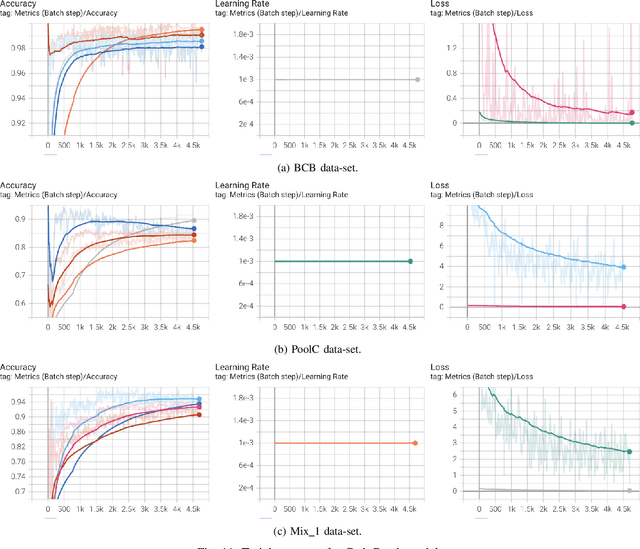
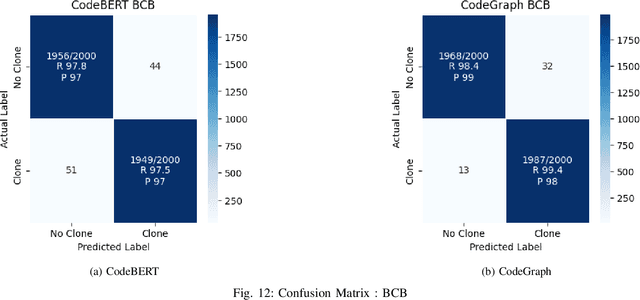
Abstract:Source code clone detection is the task of finding code fragments that have the same or similar functionality, but may differ in syntax or structure. This task is important for software maintenance, reuse, and quality assurance (Roy et al. 2009). However, code clone detection is challenging, as source code can be written in different languages, domains, and styles. In this paper, we argue that source code is inherently a graph, not a sequence, and that graph-based methods are more suitable for code clone detection than sequence-based methods. We compare the performance of two state-of-the-art models: CodeBERT (Feng et al. 2020), a sequence-based model, and CodeGraph (Yu et al. 2023), a graph-based model, on two benchmark data-sets: BCB (Svajlenko et al. 2014) and PoolC (PoolC no date). We show that CodeGraph outperforms CodeBERT on both data-sets, especially on cross-lingual code clones. To the best of our knowledge, this is the first work to demonstrate the superiority of graph-based methods over sequence-based methods on cross-lingual code clone detection.
Controlled Text Generation using T5 based Encoder-Decoder Soft Prompt Tuning and Analysis of the Utility of Generated Text in AI
Dec 06, 2022



Abstract:Controlled text generation is a very important task in the arena of natural language processing due to its promising applications. In order to achieve this task we mainly introduce the novel soft prompt tuning method of using soft prompts at both encoder and decoder levels together in a T5 model and investigate the performance as the behaviour of an additional soft prompt related to the decoder of a T5 model in controlled text generation remained unexplored. Then we also investigate the feasibility of steering the output of this extended soft prompted T5 model at decoder level and finally analyse the utility of generated text to be used in AI related tasks such as training AI models with an interpretability analysis of the classifier trained with synthetic text, as there is a lack of proper analysis of methodologies in generating properly labelled data to be utilized in AI tasks. Through the performed in-depth intrinsic and extrinsic evaluations of this generation model along with the artificially generated data, we found that this model produced better results compared to the T5 model with a single soft prompt at encoder level and the sentiment classifier trained using this artificially generated data can produce comparable classification results to the results of a classifier trained with real labelled data and also the classifier decision is interpretable with respect to the input text content.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge