Richard JB Dobson
Developing and Evaluating an AI-Assisted Prediction Model for Unplanned Intensive Care Admissions following Elective Neurosurgery using Natural Language Processing within an Electronic Healthcare Record System
Mar 13, 2025Abstract:Introduction: Timely care in a specialised neuro-intensive therapy unit (ITU) reduces mortality and hospital stays, with planned admissions being safer than unplanned ones. However, post-operative care decisions remain subjective. This study used artificial intelligence (AI), specifically natural language processing (NLP) to analyse electronic health records (EHRs) and predict ITU admissions for elective surgery patients. Methods: This study analysed the EHRs of elective neurosurgery patients from University College London Hospital (UCLH) using NLP. Patients were categorised into planned high dependency unit (HDU) or ITU admission; unplanned HDU or ITU admission; or ward / overnight recovery (ONR). The Medical Concept Annotation Tool (MedCAT) was used to identify SNOMED-CT concepts within the clinical notes. We then explored the utility of these identified concepts for a range of AI algorithms trained to predict ITU admission. Results: The CogStack-MedCAT NLP model, initially trained on hospital-wide EHRs, underwent two refinements: first with data from patients with Normal Pressure Hydrocephalus (NPH) and then with data from Vestibular Schwannoma (VS) patients, achieving a concept detection F1-score of 0.93. This refined model was then used to extract concepts from EHR notes of 2,268 eligible neurosurgical patients. We integrated the extracted concepts into AI models, including a decision tree model and a neural time-series model. Using the simpler decision tree model, we achieved a recall of 0.87 (CI 0.82 - 0.91) for ITU admissions, reducing the proportion of unplanned ITU cases missed by human experts from 36% to 4%. Conclusion: The NLP model, refined for accuracy, has proven its efficiency in extracting relevant concepts, providing a reliable basis for predictive AI models to use in clinically valid applications.
Unveiling the Secrets: How Masking Strategies Shape Time Series Imputation
May 26, 2024Abstract:In this study, we explore the impact of different masking strategies on time series imputation models. We evaluate the effects of pre-masking versus in-mini-batch masking, normalization timing, and the choice between augmenting and overlaying artificial missingness. Using three diverse datasets, we benchmark eleven imputation models with different missing rates. Our results demonstrate that masking strategies significantly influence imputation accuracy, revealing that more sophisticated and data-driven masking designs are essential for robust model evaluation. We advocate for refined experimental designs and comprehensive disclosureto better simulate real-world patterns, enhancing the practical applicability of imputation models.
Identifying depression-related topics in smartphone-collected free-response speech recordings using an automatic speech recognition system and a deep learning topic model
Sep 05, 2023



Abstract:Language use has been shown to correlate with depression, but large-scale validation is needed. Traditional methods like clinic studies are expensive. So, natural language processing has been employed on social media to predict depression, but limitations remain-lack of validated labels, biased user samples, and no context. Our study identified 29 topics in 3919 smartphone-collected speech recordings from 265 participants using the Whisper tool and BERTopic model. Six topics with a median PHQ-8 greater than or equal to 10 were regarded as risk topics for depression: No Expectations, Sleep, Mental Therapy, Haircut, Studying, and Coursework. To elucidate the topic emergence and associations with depression, we compared behavioral (from wearables) and linguistic characteristics across identified topics. The correlation between topic shifts and changes in depression severity over time was also investigated, indicating the importance of longitudinally monitoring language use. We also tested the BERTopic model on a similar smaller dataset (356 speech recordings from 57 participants), obtaining some consistent results. In summary, our findings demonstrate specific speech topics may indicate depression severity. The presented data-driven workflow provides a practical approach to collecting and analyzing large-scale speech data from real-world settings for digital health research.
Disease Insight through Digital Biomarkers Developed by Remotely Collected Wearables and Smartphone Data
Aug 03, 2023



Abstract:Digital Biomarkers and remote patient monitoring can provide valuable and timely insights into how a patient is coping with their condition (disease progression, treatment response, etc.), complementing treatment in traditional healthcare settings.Smartphones with embedded and connected sensors have immense potential for improving healthcare through various apps and mHealth (mobile health) platforms. This capability could enable the development of reliable digital biomarkers from long-term longitudinal data collected remotely from patients. We built an open-source platform, RADAR-base, to support large-scale data collection in remote monitoring studies. RADAR-base is a modern remote data collection platform built around Confluent's Apache Kafka, to support scalability, extensibility, security, privacy and quality of data. It provides support for study design and set-up, active (eg PROMs) and passive (eg. phone sensors, wearable devices and IoT) remote data collection capabilities with feature generation (eg. behavioural, environmental and physiological markers). The backend enables secure data transmission, and scalable solutions for data storage, management and data access. The platform has successfully collected longitudinal data for various cohorts in a number of disease areas including Multiple Sclerosis, Depression, Epilepsy, ADHD, Alzheimer, Autism and Lung diseases. Digital biomarkers developed through collected data are providing useful insights into different diseases. RADAR-base provides a modern open-source, community-driven solution for remote monitoring, data collection, and digital phenotyping of physical and mental health diseases. Clinicians can use digital biomarkers to augment their decision making for the prevention, personalisation and early intervention of disease.
Estimating Redundancy in Clinical Text
May 25, 2021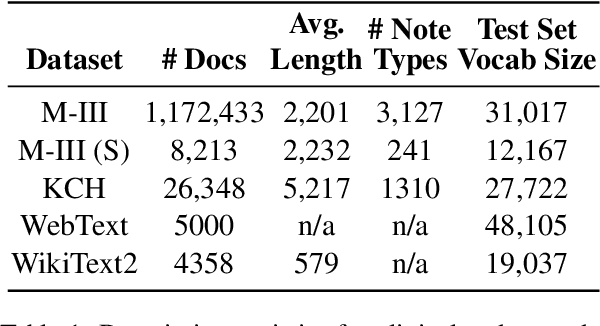
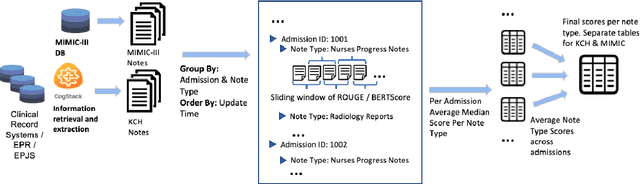
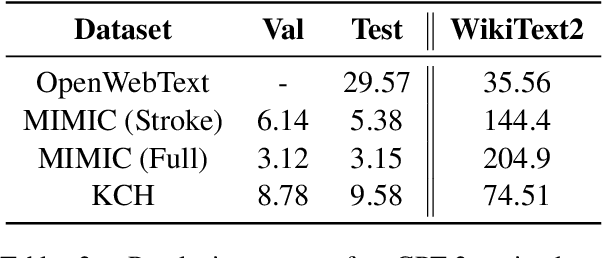
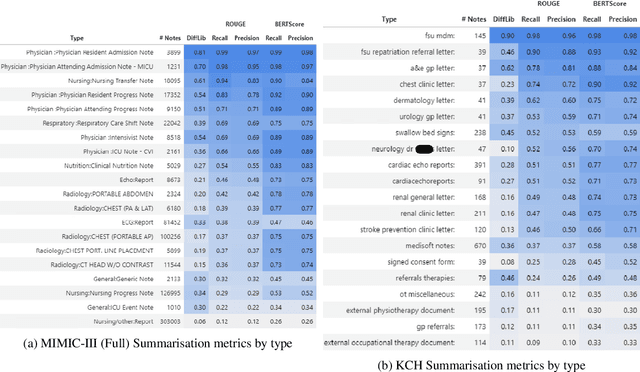
Abstract:The current mode of use of Electronic Health Record (EHR) elicits text redundancy. Clinicians often populate new documents by duplicating existing notes, then updating accordingly. Data duplication can lead to a propagation of errors, inconsistencies and misreporting of care. Therefore, quantifying information redundancy can play an essential role in evaluating innovations that operate on clinical narratives. This work is a quantitative examination of information redundancy in EHR notes. We present and evaluate two strategies to measure redundancy: an information-theoretic approach and a lexicosyntactic and semantic model. We evaluate the measures by training large Transformer-based language models using clinical text from a large openly available US-based ICU dataset and a large multi-site UK based Trust. By comparing the information-theoretic content of the trained models with open-domain language models, the language models trained using clinical text have shown ~1.5x to ~3x less efficient than open-domain corpora. Manual evaluation shows a high correlation with lexicosyntactic and semantic redundancy, with averages ~43 to ~65%.
Predicting Depressive Symptom Severity through Individuals' Nearby Bluetooth Devices Count Data Collected by Mobile Phones: A Preliminary Longitudinal Study
Apr 26, 2021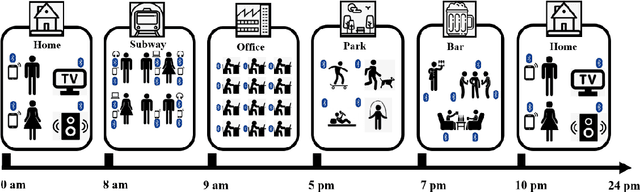
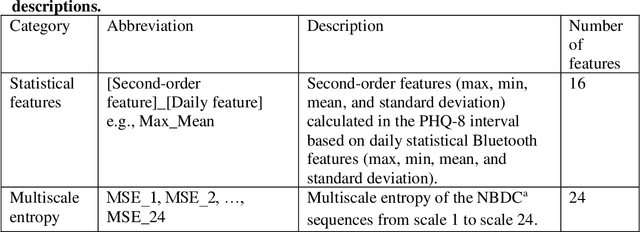
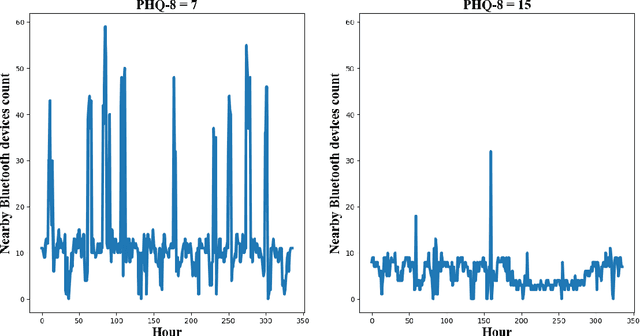
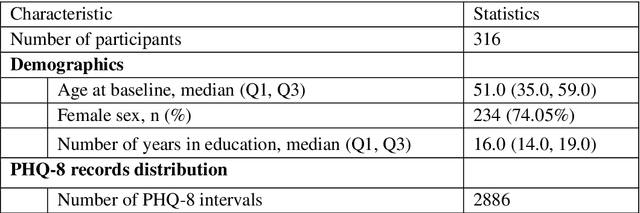
Abstract:The Bluetooth sensor embedded in mobile phones provides an unobtrusive, continuous, and cost-efficient means to capture individuals' proximity information, such as the nearby Bluetooth devices count (NBDC). The continuous NBDC data can partially reflect individuals' behaviors and status, such as social connections and interactions, working status, mobility, and social isolation and loneliness, which were found to be significantly associated with depression by previous survey-based studies. This paper aims to explore the NBDC data's value in predicting depressive symptom severity as measured via the 8-item Patient Health Questionnaire (PHQ-8). The data used in this paper included 2,886 bi-weekly PHQ-8 records collected from 316 participants recruited from three study sites in the Netherlands, Spain, and the UK as part of the EU RADAR-CNS study. From the NBDC data two weeks prior to each PHQ-8 score, we extracted 49 Bluetooth features, including statistical features and nonlinear features for measuring periodicity and regularity of individuals' life rhythms. Linear mixed-effect models were used to explore associations between Bluetooth features and the PHQ-8 score. We then applied hierarchical Bayesian linear regression models to predict the PHQ-8 score from the extracted Bluetooth features. A number of significant associations were found between Bluetooth features and depressive symptom severity. Compared with commonly used machine learning models, the proposed hierarchical Bayesian linear regression model achieved the best prediction metrics, R2= 0.526, and root mean squared error (RMSE) of 3.891. Bluetooth features can explain an extra 18.8% of the variance in the PHQ-8 score relative to the baseline model without Bluetooth features (R2=0.338, RMSE = 4.547).
Fitbeat: COVID-19 Estimation based on Wristband Heart Rate
Apr 19, 2021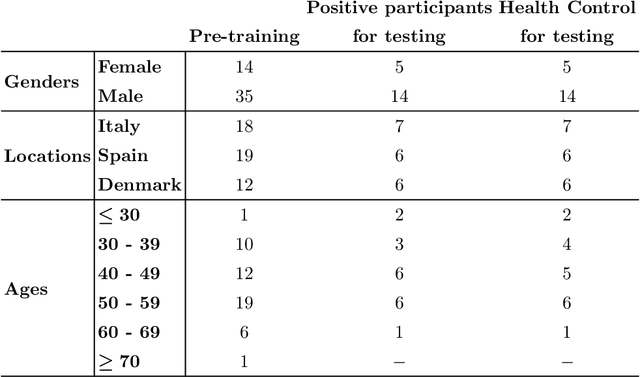

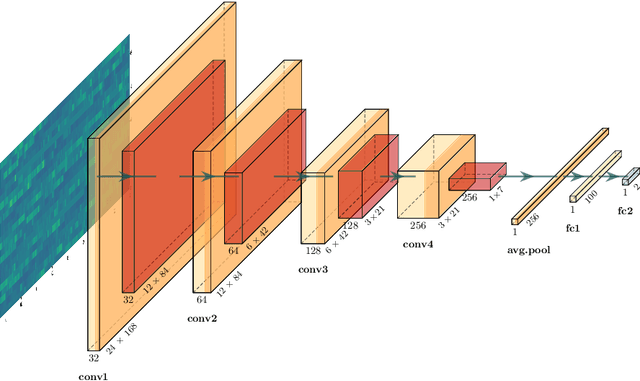
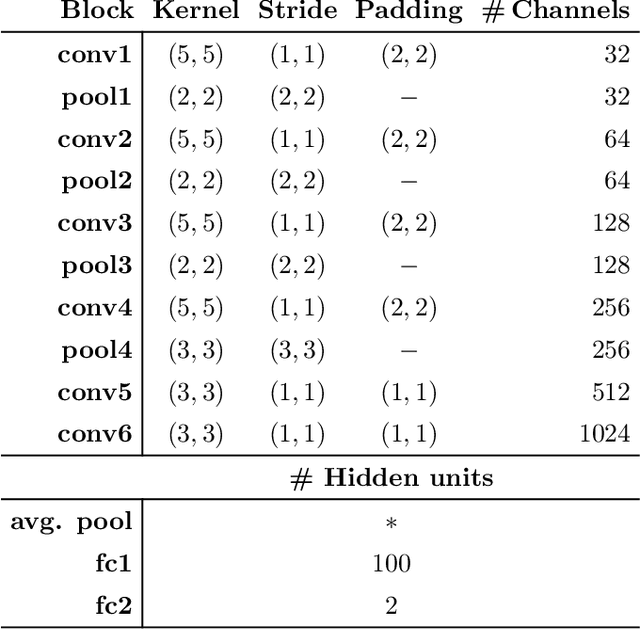
Abstract:This study investigates the potential of deep learning methods to identify individuals with suspected COVID-19 infection using remotely collected heart-rate data. The study utilises data from the ongoing EU IMI RADAR-CNS research project that is investigating the feasibility of wearable devices and smart phones to monitor individuals with multiple sclerosis (MS), depression or epilepsy. Aspart of the project protocol, heart-rate data was collected from participants using a Fitbit wristband. The presence of COVID-19 in the cohort in this work was either confirmed through a positive swab test, or inferred through the self-reporting of a combination of symptoms including fever, respiratory symptoms, loss of smell or taste, tiredness and gastrointestinal symptoms. Experimental results indicate that our proposed contrastive convolutional auto-encoder (contrastive CAE), i. e., a combined architecture of an auto-encoder and contrastive loss, outperforms a conventional convolutional neural network (CNN), as well as a convolutional auto-encoder (CAE) without using contrastive loss. Our final contrastive CAE achieves 95.3% unweighted average recall, 86.4% precision, anF1 measure of 88.2%, a sensitivity of 100% and a specificity of 90.6% on a testset of 19 participants with MS who reported symptoms of COVID-19. Each of these participants was paired with a participant with MS with no COVID-19 symptoms.
A Knowledge Distillation Ensemble Framework for Predicting Short and Long-term Hospitalisation Outcomes from Electronic Health Records Data
Nov 18, 2020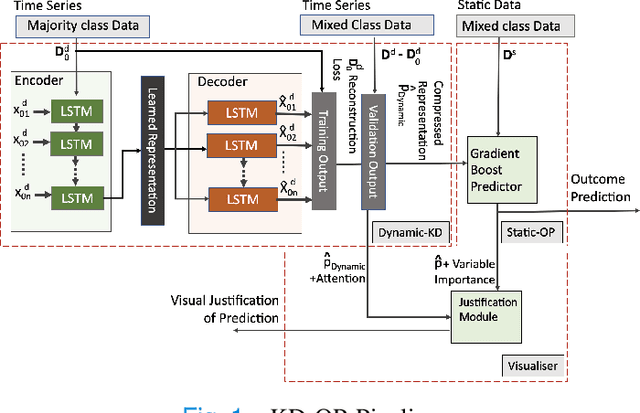
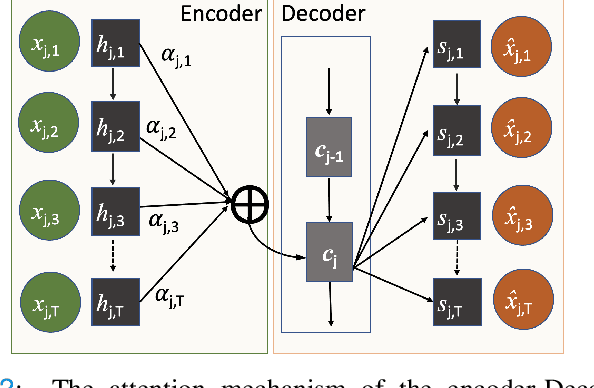
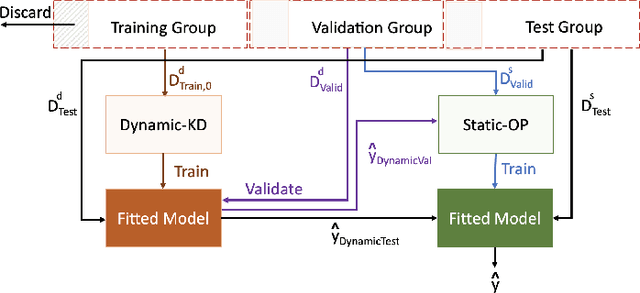
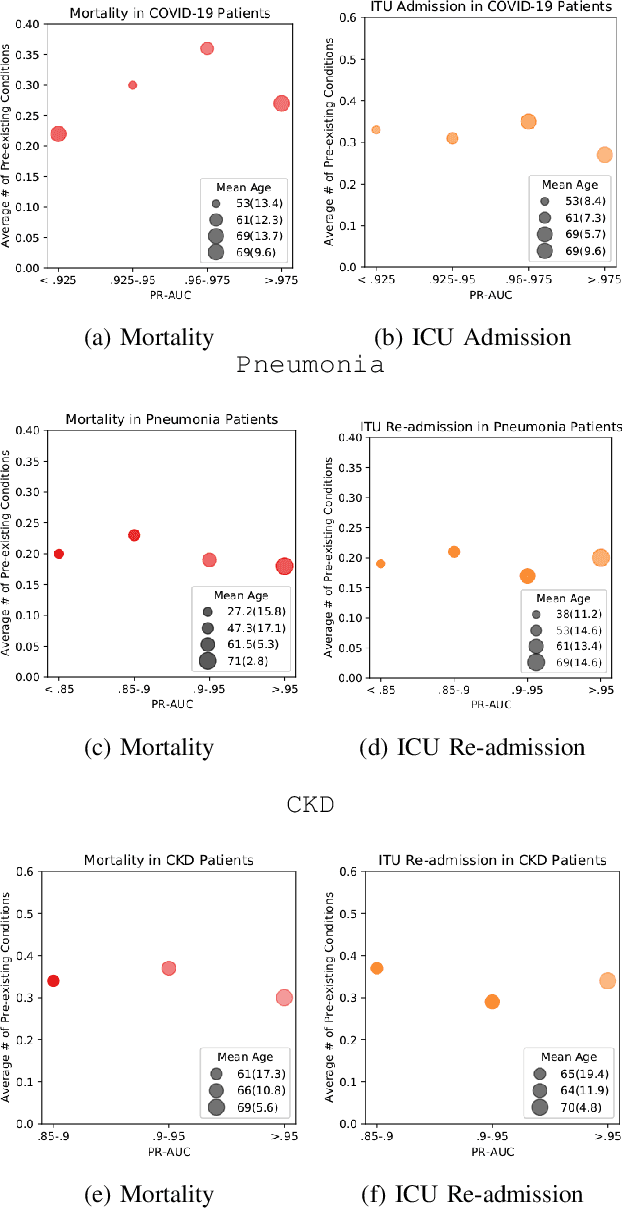
Abstract:The ability to perform accurate prognosis of patients is crucial for proactive clinical decision making, informed resource management and personalised care. Existing outcome prediction models suffer from a low recall of infrequent positive outcomes. We present a highly-scalable and robust machine learning framework to automatically predict adversity represented by mortality and ICU admission from time-series vital signs and laboratory results obtained within the first 24 hours of hospital admission. The stacked platform comprises two components: a) an unsupervised LSTM Autoencoder that learns an optimal representation of the time-series, using it to differentiate the less frequent patterns which conclude with an adverse event from the majority patterns that do not, and b) a gradient boosting model, which relies on the constructed representation to refine prediction, incorporating static features of demographics, admission details and clinical summaries. The model is used to assess a patient's risk of adversity over time and provides visual justifications of its prediction based on the patient's static features and dynamic signals. Results of three case studies for predicting mortality and ICU admission show that the model outperforms all existing outcome prediction models, achieving PR-AUC of 0.93 (95$%$ CI: 0.878 - 0.969) in predicting mortality in ICU and general ward settings and 0.987 (95$%$ CI: 0.985-0.995) in predicting ICU admission.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020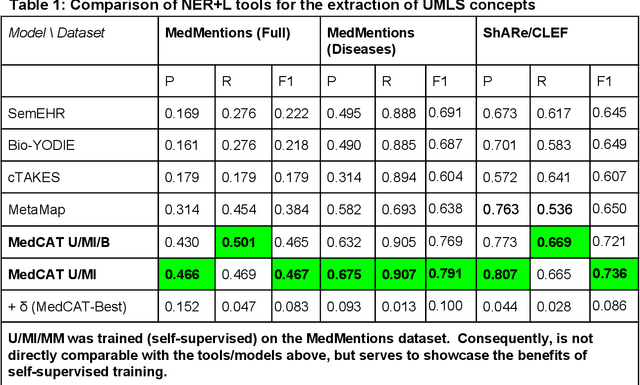
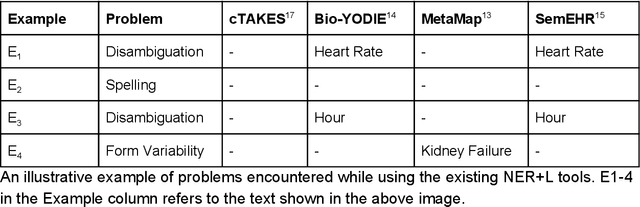
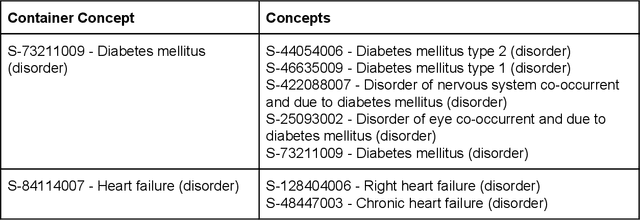
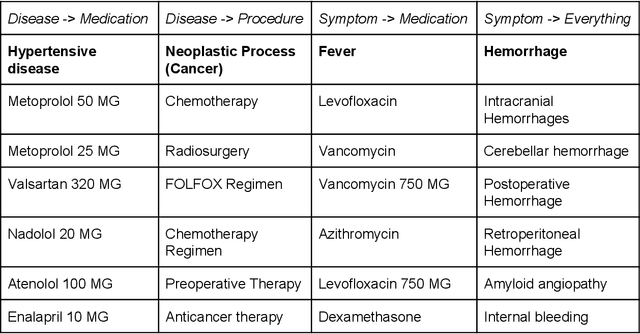
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
Experimental Evaluation and Development of a Silver-Standard for the MIMIC-III Clinical Coding Dataset
Jun 12, 2020



Abstract:Clinical coding is currently a labour-intensive, error-prone, but critical administrative process whereby hospital patient episodes are manually assigned codes by qualified staff from large, standardised taxonomic hierarchies of codes. Automating clinical coding has a long history in NLP research and has recently seen novel developments setting new state of the art results. A popular dataset used in this task is MIMIC-III, a large intensive care database that includes clinical free text notes and associated codes. We argue for the reconsideration of the validity MIMIC-III's assigned codes that are often treated as gold-standard, especially when MIMIC-III has not undergone secondary validation. This work presents an open-source, reproducible experimental methodology for assessing the validity of codes derived from EHR discharge summaries. We exemplify the methodology with MIMIC-III discharge summaries and show the most frequently assigned codes in MIMIC-III are under-coded up to 35%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge