Thomas Searle
RelCAT: Advancing Extraction of Clinical Inter-Entity Relationships from Unstructured Electronic Health Records
Jan 27, 2025Abstract:This study introduces RelCAT (Relation Concept Annotation Toolkit), an interactive tool, library, and workflow designed to classify relations between entities extracted from clinical narratives. Building upon the CogStack MedCAT framework, RelCAT addresses the challenge of capturing complete clinical relations dispersed within text. The toolkit implements state-of-the-art machine learning models such as BERT and Llama along with proven evaluation and training methods. We demonstrate a dataset annotation tool (built within MedCATTrainer), model training, and evaluate our methodology on both openly available gold-standard and real-world UK National Health Service (NHS) hospital clinical datasets. We perform extensive experimentation and a comparative analysis of the various publicly available models with varied approaches selected for model fine-tuning. Finally, we achieve macro F1-scores of 0.977 on the gold-standard n2c2, surpassing the previous state-of-the-art performance, and achieve performance of >=0.93 F1 on our NHS gathered datasets.
Improving Extraction of Clinical Event Contextual Properties from Electronic Health Records: A Comparative Study
Aug 30, 2024
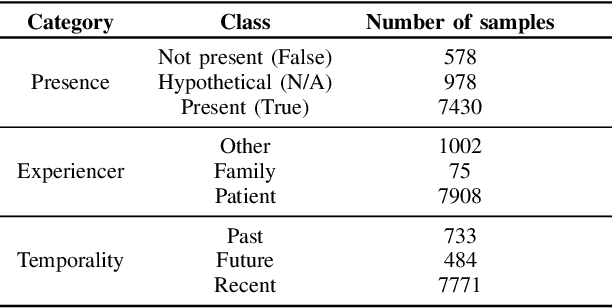
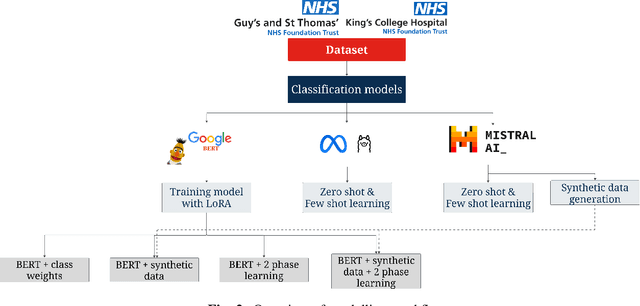
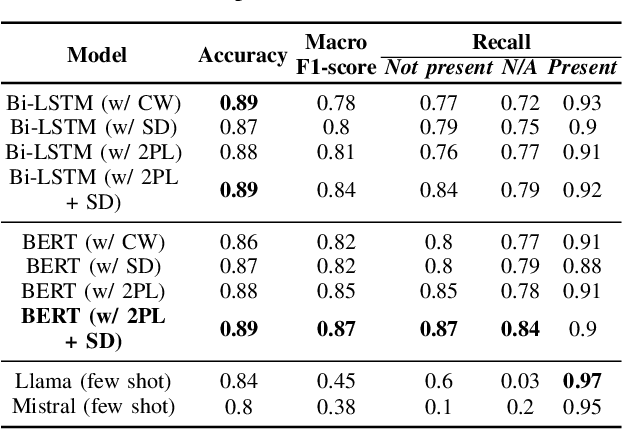
Abstract:Electronic Health Records are large repositories of valuable clinical data, with a significant portion stored in unstructured text format. This textual data includes clinical events (e.g., disorders, symptoms, findings, medications and procedures) in context that if extracted accurately at scale can unlock valuable downstream applications such as disease prediction. Using an existing Named Entity Recognition and Linking methodology, MedCAT, these identified concepts need to be further classified (contextualised) for their relevance to the patient, and their temporal and negated status for example, to be useful downstream. This study performs a comparative analysis of various natural language models for medical text classification. Extensive experimentation reveals the effectiveness of transformer-based language models, particularly BERT. When combined with class imbalance mitigation techniques, BERT outperforms Bi-LSTM models by up to 28% and the baseline BERT model by up to 16% for recall of the minority classes. The method has been implemented as part of CogStack/MedCAT framework and made available to the community for further research.
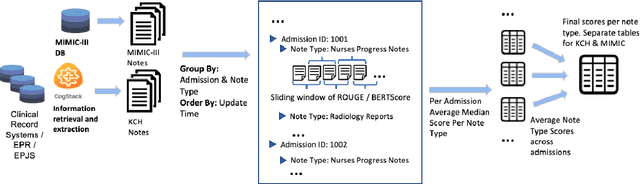
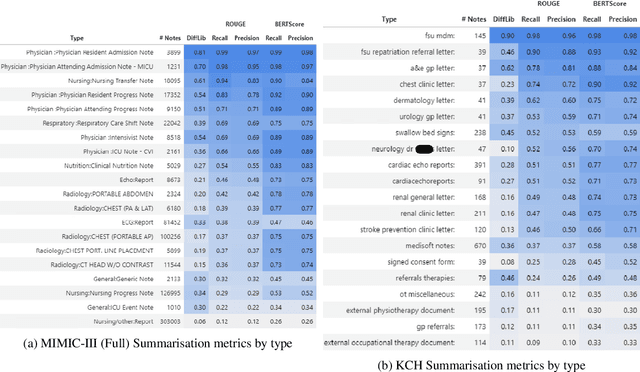
Summarisation of Electronic Health Records with Clinical Concept Guidance
Nov 14, 2022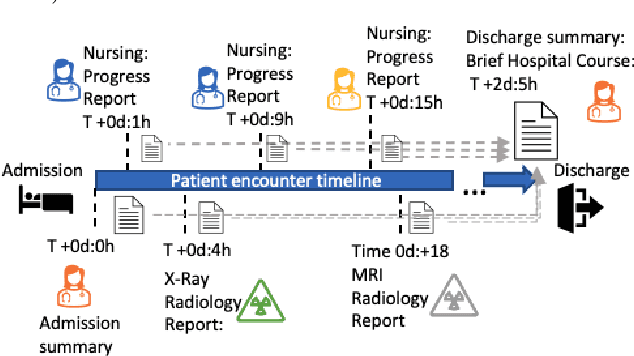
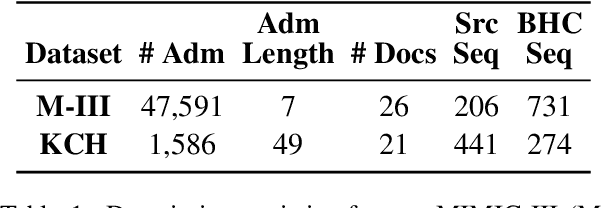
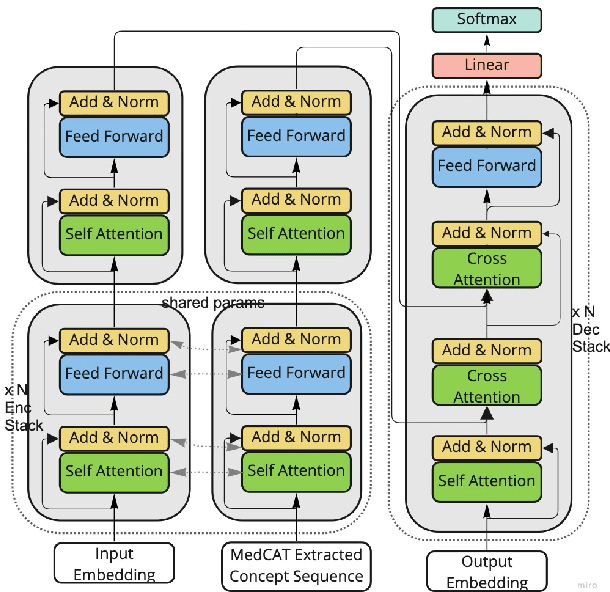
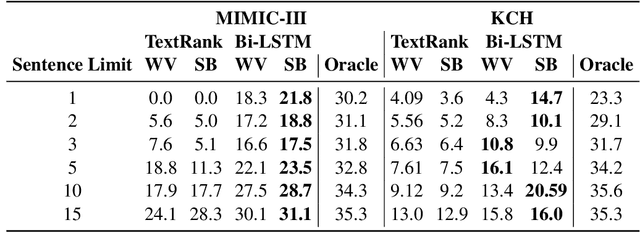
Abstract:Brief Hospital Course (BHC) summaries are succinct summaries of an entire hospital encounter, embedded within discharge summaries, written by senior clinicians responsible for the overall care of a patient. Methods to automatically produce summaries from inpatient documentation would be invaluable in reducing clinician manual burden of summarising documents under high time-pressure to admit and discharge patients. Automatically producing these summaries from the inpatient course, is a complex, multi-document summarisation task, as source notes are written from various perspectives (e.g. nursing, doctor, radiology), during the course of the hospitalisation. We demonstrate a range of methods for BHC summarisation demonstrating the performance of deep learning summarisation models across extractive and abstractive summarisation scenarios. We also test a novel ensemble extractive and abstractive summarisation model that incorporates a medical concept ontology (SNOMED) as a clinical guidance signal and shows superior performance in 2 real-world clinical data sets.
Estimating Redundancy in Clinical Text
May 25, 2021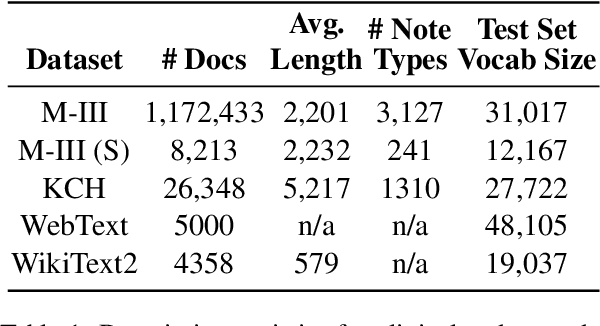
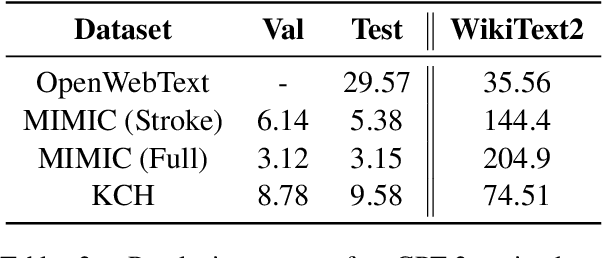
Abstract:The current mode of use of Electronic Health Record (EHR) elicits text redundancy. Clinicians often populate new documents by duplicating existing notes, then updating accordingly. Data duplication can lead to a propagation of errors, inconsistencies and misreporting of care. Therefore, quantifying information redundancy can play an essential role in evaluating innovations that operate on clinical narratives. This work is a quantitative examination of information redundancy in EHR notes. We present and evaluate two strategies to measure redundancy: an information-theoretic approach and a lexicosyntactic and semantic model. We evaluate the measures by training large Transformer-based language models using clinical text from a large openly available US-based ICU dataset and a large multi-site UK based Trust. By comparing the information-theoretic content of the trained models with open-domain language models, the language models trained using clinical text have shown ~1.5x to ~3x less efficient than open-domain corpora. Manual evaluation shows a high correlation with lexicosyntactic and semantic redundancy, with averages ~43 to ~65%.
A Knowledge Distillation Ensemble Framework for Predicting Short and Long-term Hospitalisation Outcomes from Electronic Health Records Data
Nov 18, 2020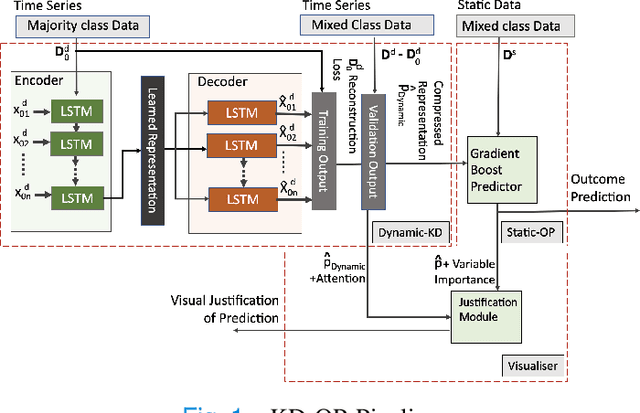
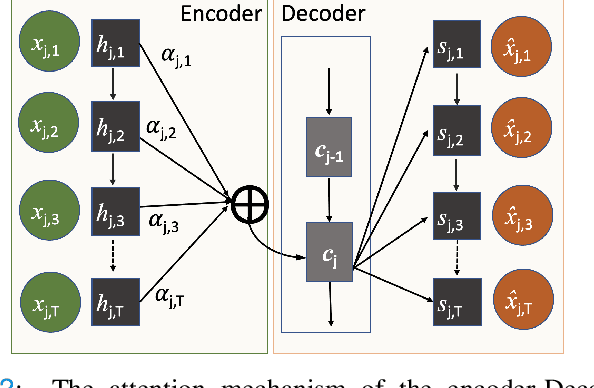
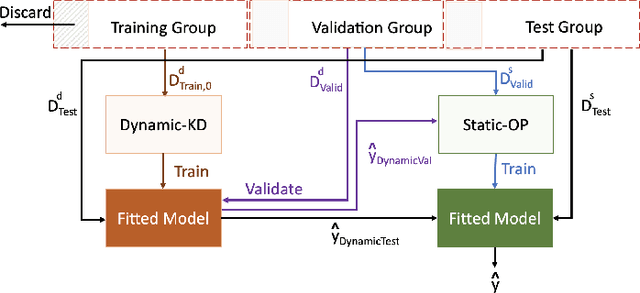
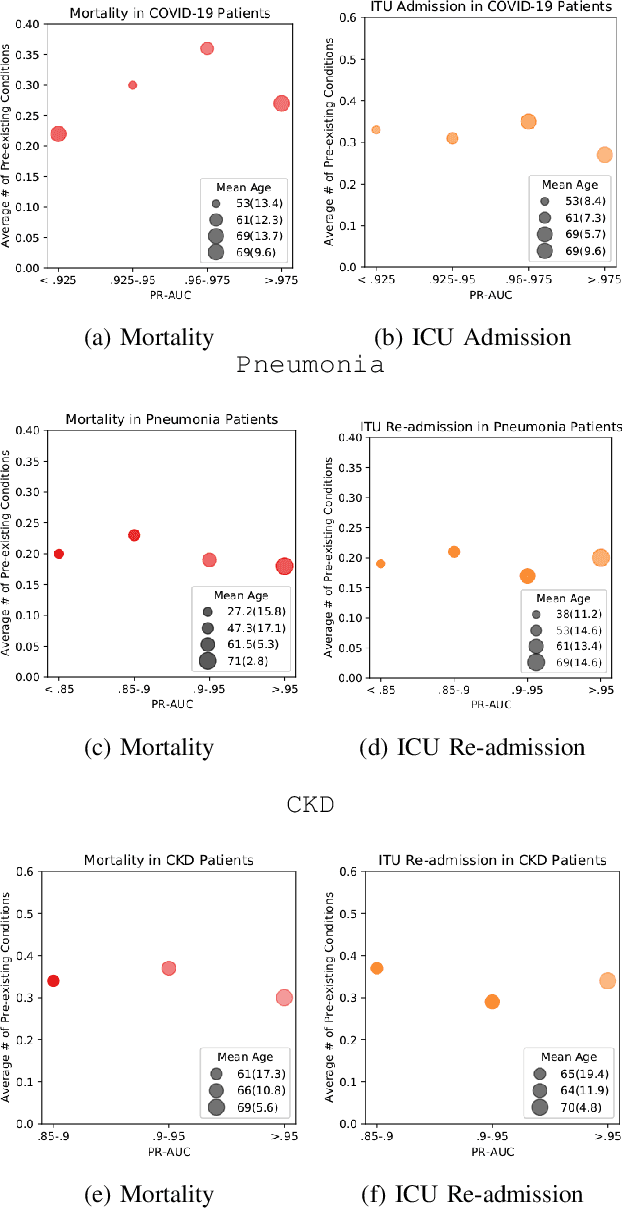
Abstract:The ability to perform accurate prognosis of patients is crucial for proactive clinical decision making, informed resource management and personalised care. Existing outcome prediction models suffer from a low recall of infrequent positive outcomes. We present a highly-scalable and robust machine learning framework to automatically predict adversity represented by mortality and ICU admission from time-series vital signs and laboratory results obtained within the first 24 hours of hospital admission. The stacked platform comprises two components: a) an unsupervised LSTM Autoencoder that learns an optimal representation of the time-series, using it to differentiate the less frequent patterns which conclude with an adverse event from the majority patterns that do not, and b) a gradient boosting model, which relies on the constructed representation to refine prediction, incorporating static features of demographics, admission details and clinical summaries. The model is used to assess a patient's risk of adversity over time and provides visual justifications of its prediction based on the patient's static features and dynamic signals. Results of three case studies for predicting mortality and ICU admission show that the model outperforms all existing outcome prediction models, achieving PR-AUC of 0.93 (95$%$ CI: 0.878 - 0.969) in predicting mortality in ICU and general ward settings and 0.987 (95$%$ CI: 0.985-0.995) in predicting ICU admission.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020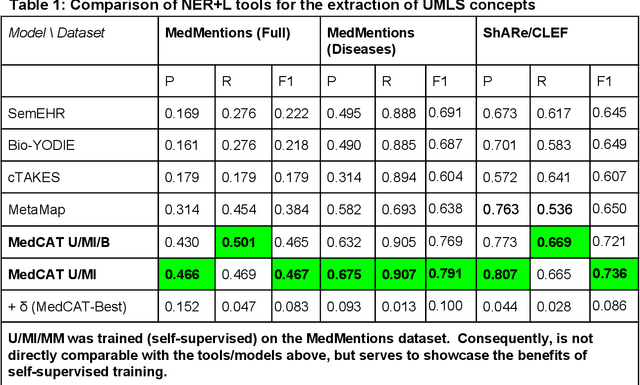
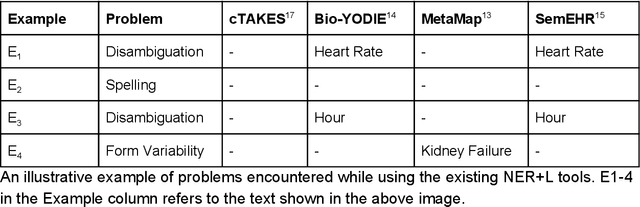
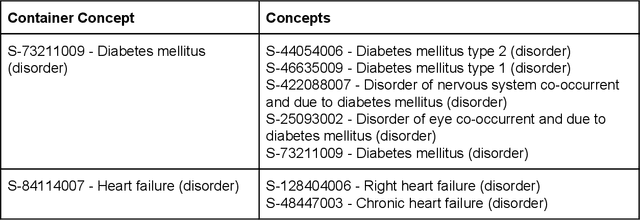
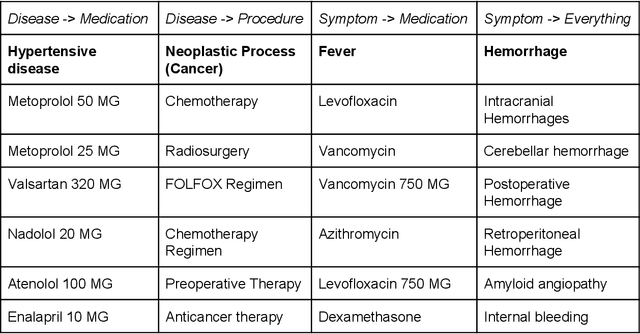
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
Comparing Natural Language Processing Techniques for Alzheimer's Dementia Prediction in Spontaneous Speech
Jun 12, 2020



Abstract:Alzheimer's Dementia (AD) is an incurable, debilitating, and progressive neurodegenerative condition that affects cognitive function. Early diagnosis is important as therapeutics can delay progression and give those diagnosed vital time. Developing models that analyse spontaneous speech could eventually provide an efficient diagnostic modality for earlier diagnosis of AD. The Alzheimer's Dementia Recognition through Spontaneous Speech task offers acoustically pre-processed and balanced datasets for the classification and prediction of AD and associated phenotypes through the modelling of spontaneous speech. We exclusively analyse the supplied textual transcripts of the spontaneous speech dataset, building and comparing performance across numerous models for the classification of AD vs controls and the prediction of Mental Mini State Exam scores. We rigorously train and evaluate Support Vector Machines (SVMs), Gradient Boosting Decision Trees (GBDT), and Conditional Random Fields (CRFs) alongside deep learning Transformer based models. We find our top performing models to be a simple Term Frequency-Inverse Document Frequency (TF-IDF) vectoriser as input into a SVM model and a pre-trained Transformer based model `DistilBERT' when used as an embedding layer into simple linear models. We demonstrate test set scores of 0.81-0.82 across classification metrics and a RMSE of 4.58.
Experimental Evaluation and Development of a Silver-Standard for the MIMIC-III Clinical Coding Dataset
Jun 12, 2020



Abstract:Clinical coding is currently a labour-intensive, error-prone, but critical administrative process whereby hospital patient episodes are manually assigned codes by qualified staff from large, standardised taxonomic hierarchies of codes. Automating clinical coding has a long history in NLP research and has recently seen novel developments setting new state of the art results. A popular dataset used in this task is MIMIC-III, a large intensive care database that includes clinical free text notes and associated codes. We argue for the reconsideration of the validity MIMIC-III's assigned codes that are often treated as gold-standard, especially when MIMIC-III has not undergone secondary validation. This work presents an open-source, reproducible experimental methodology for assessing the validity of codes derived from EHR discharge summaries. We exemplify the methodology with MIMIC-III discharge summaries and show the most frequently assigned codes in MIMIC-III are under-coded up to 35%.
MedCATTrainer: A Biomedical Free Text Annotation Interface with Active Learning and Research Use Case Specific Customisation
Jul 16, 2019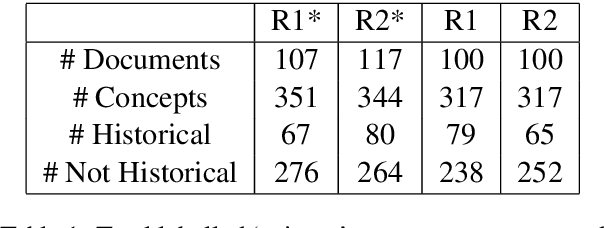
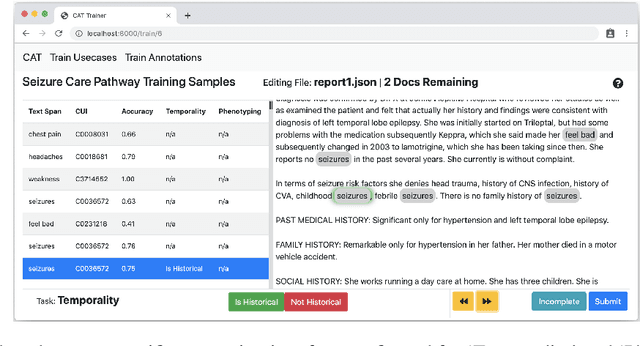
Abstract:We present MedCATTrainer an interface for building, improving and customising a given Named Entity Recognition and Linking (NER+L) model for biomedical domain text. NER+L is often used as a first step in deriving value from clinical text. Collecting labelled data for training models is difficult due to the need for specialist domain knowledge. MedCATTrainer offers an interactive web-interface to inspect and improve recognised entities from an underlying NER+L model via active learning. Secondary use of data for clinical research often has task and context specific criteria. MedCATTrainer provides a further interface to define and collect supervised learning training data for researcher specific use cases. Initial results suggest our approach allows for efficient and accurate collection of research use case specific training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge