Daniel Bean
Large Language Models for Medical Forecasting -- Foresight 2
Dec 14, 2024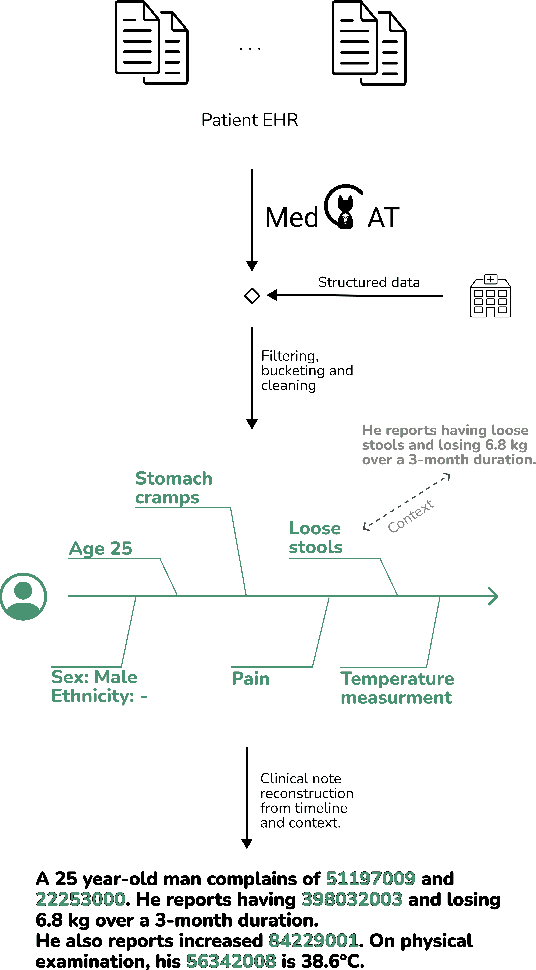
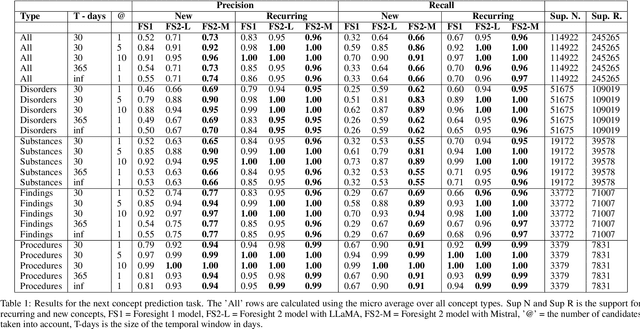
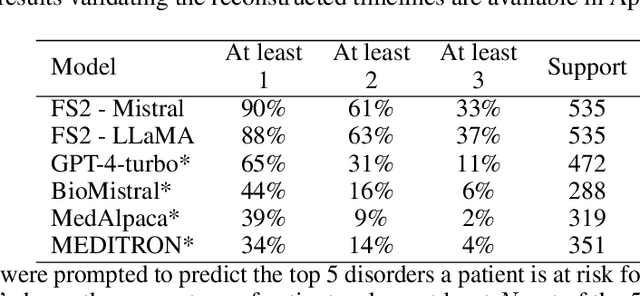
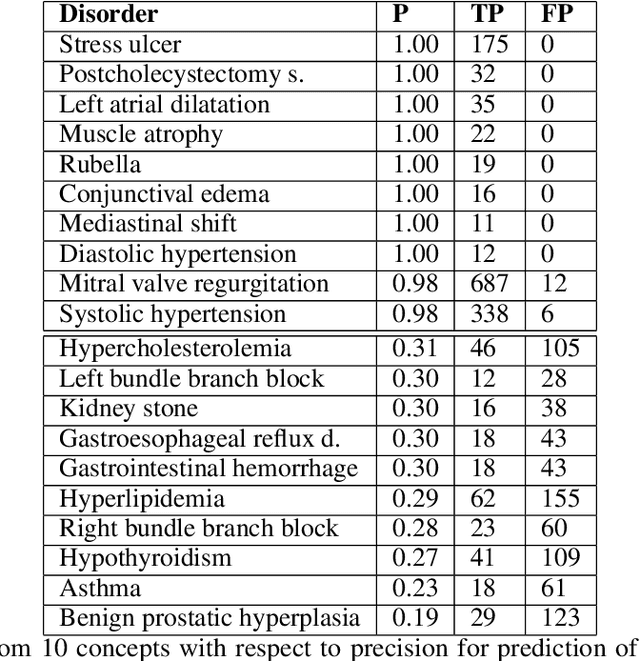
Abstract:Foresight 2 (FS2) is a large language model fine-tuned on hospital data for modelling patient timelines (GitHub 'removed for anon'). It can understand patients' clinical notes and predict SNOMED codes for a wide range of biomedical use cases, including diagnosis suggestions, risk forecasting, and procedure and medication recommendations. FS2 is trained on the free text portion of the MIMIC-III dataset, firstly through extracting biomedical concepts and then creating contextualised patient timelines, upon which the model is then fine-tuned. The results show significant improvement over the previous state-of-the-art for the next new biomedical concept prediction (P/R - 0.73/0.66 vs 0.52/0.32) and a similar improvement specifically for the next new disorder prediction (P/R - 0.69/0.62 vs 0.46/0.25). Finally, on the task of risk forecast, we compare our model to GPT-4-turbo (and a range of open-source biomedical LLMs) and show that FS2 performs significantly better on such tasks (P@5 - 0.90 vs 0.65). This highlights the need to incorporate hospital data into LLMs and shows that small models outperform much larger ones when fine-tuned on high-quality, specialised data.
MedGPT: Medical Concept Prediction from Clinical Narratives
Jul 07, 2021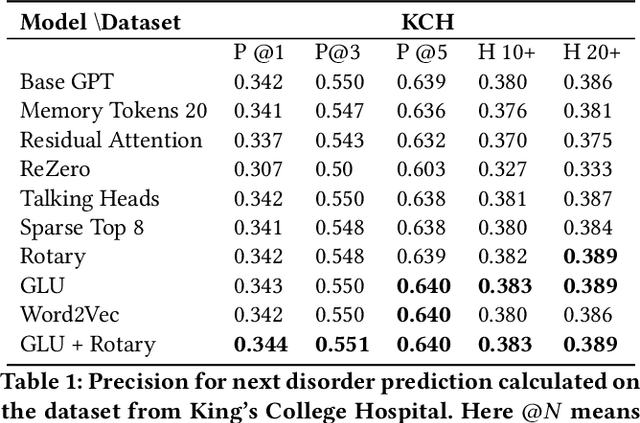
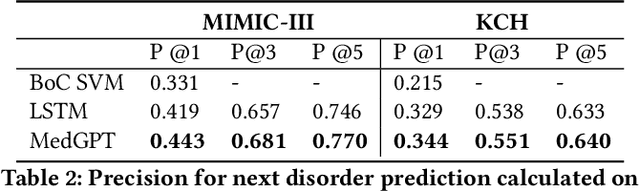
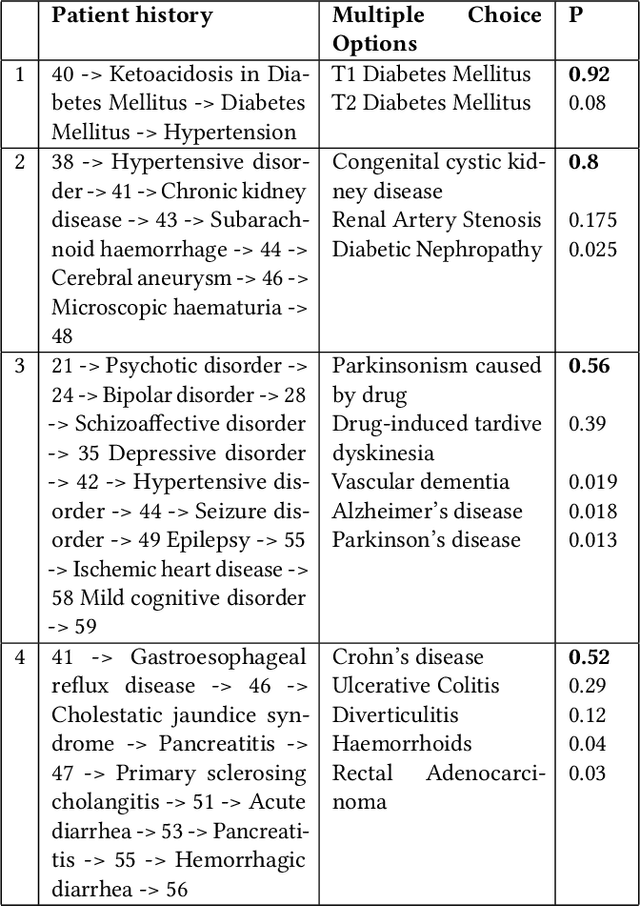
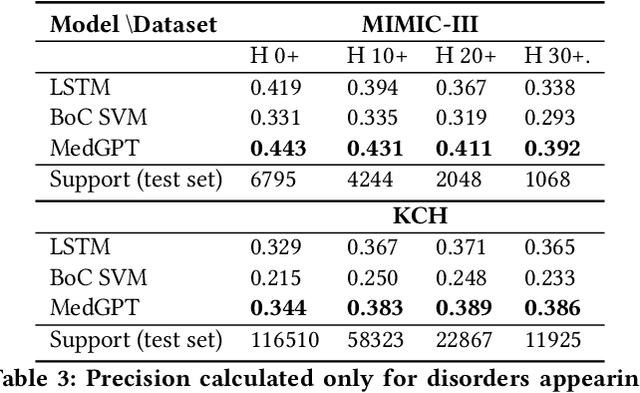
Abstract:The data available in Electronic Health Records (EHRs) provides the opportunity to transform care, and the best way to provide better care for one patient is through learning from the data available on all other patients. Temporal modelling of a patient's medical history, which takes into account the sequence of past events, can be used to predict future events such as a diagnosis of a new disorder or complication of a previous or existing disorder. While most prediction approaches use mostly the structured data in EHRs or a subset of single-domain predictions and outcomes, we present MedGPT a novel transformer-based pipeline that uses Named Entity Recognition and Linking tools (i.e. MedCAT) to structure and organize the free text portion of EHRs and anticipate a range of future medical events (initially disorders). Since a large portion of EHR data is in text form, such an approach benefits from a granular and detailed view of a patient while introducing modest additional noise. MedGPT effectively deals with the noise and the added granularity, and achieves a precision of 0.344, 0.552 and 0.640 (vs LSTM 0.329, 0.538 and 0.633) when predicting the top 1, 3 and 5 candidate future disorders on real world hospital data from King's College Hospital, London, UK (\textasciitilde600k patients). We also show that our model captures medical knowledge by testing it on an experimental medical multiple choice question answering task, and by examining the attentional focus of the model using gradient-based saliency methods.
A Knowledge Distillation Ensemble Framework for Predicting Short and Long-term Hospitalisation Outcomes from Electronic Health Records Data
Nov 18, 2020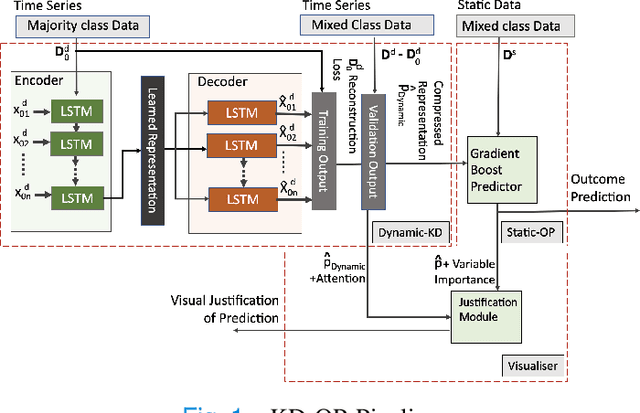
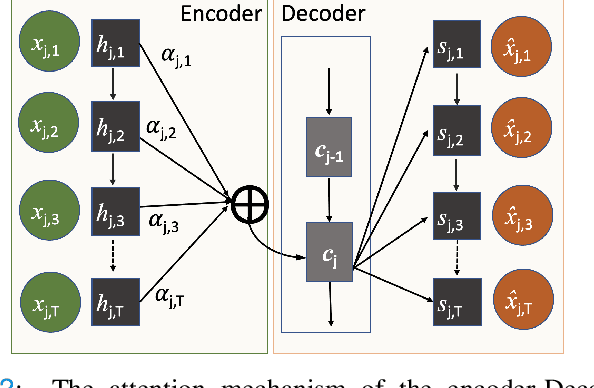
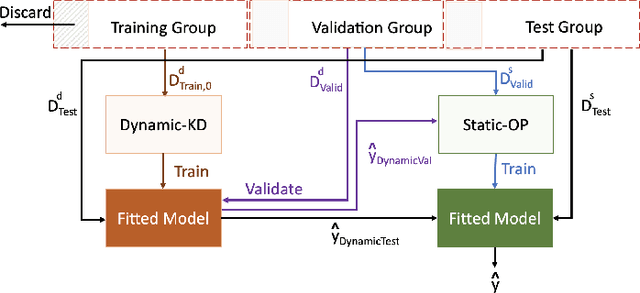
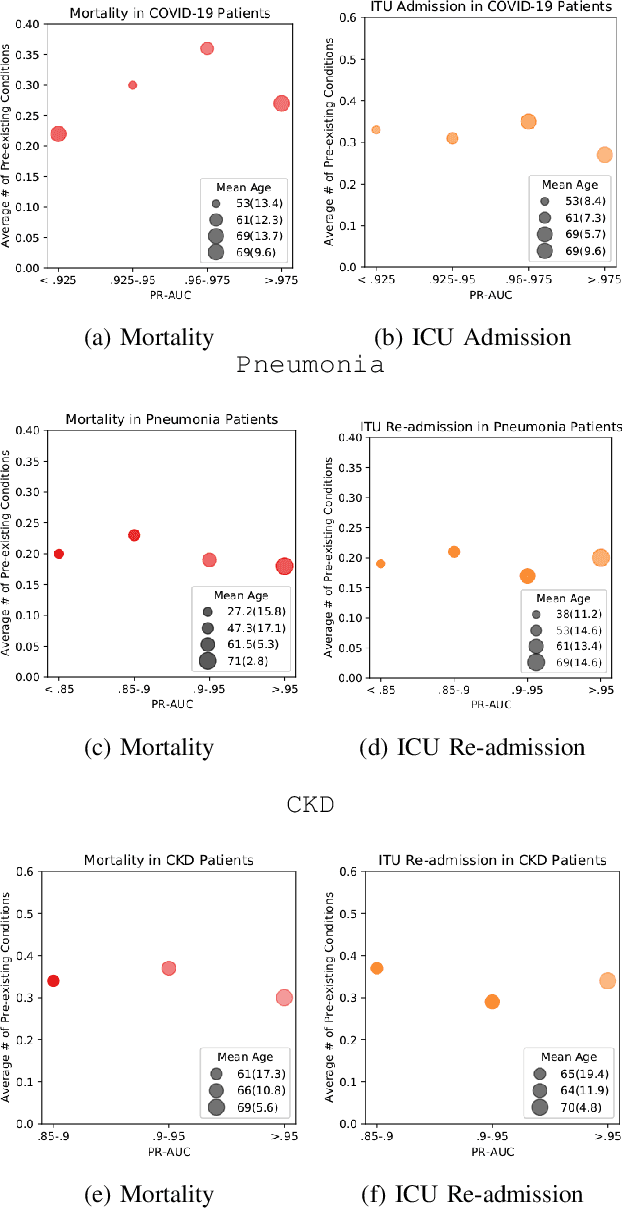
Abstract:The ability to perform accurate prognosis of patients is crucial for proactive clinical decision making, informed resource management and personalised care. Existing outcome prediction models suffer from a low recall of infrequent positive outcomes. We present a highly-scalable and robust machine learning framework to automatically predict adversity represented by mortality and ICU admission from time-series vital signs and laboratory results obtained within the first 24 hours of hospital admission. The stacked platform comprises two components: a) an unsupervised LSTM Autoencoder that learns an optimal representation of the time-series, using it to differentiate the less frequent patterns which conclude with an adverse event from the majority patterns that do not, and b) a gradient boosting model, which relies on the constructed representation to refine prediction, incorporating static features of demographics, admission details and clinical summaries. The model is used to assess a patient's risk of adversity over time and provides visual justifications of its prediction based on the patient's static features and dynamic signals. Results of three case studies for predicting mortality and ICU admission show that the model outperforms all existing outcome prediction models, achieving PR-AUC of 0.93 (95$%$ CI: 0.878 - 0.969) in predicting mortality in ICU and general ward settings and 0.987 (95$%$ CI: 0.985-0.995) in predicting ICU admission.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020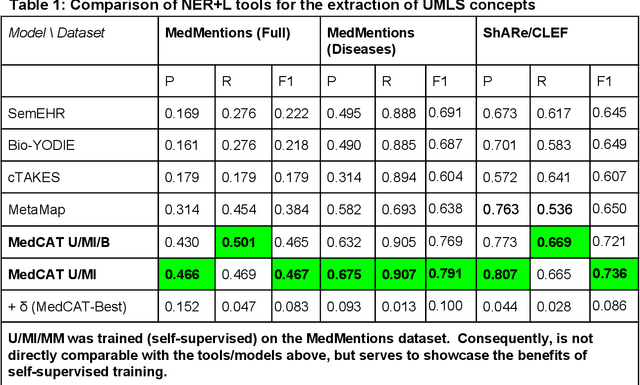
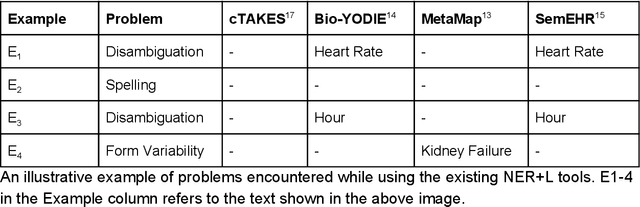
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
Comparative Analysis of Text Classification Approaches in Electronic Health Records
May 08, 2020
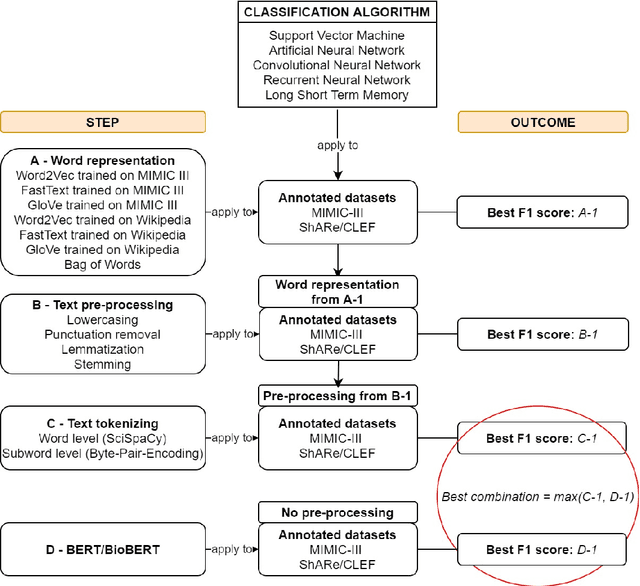

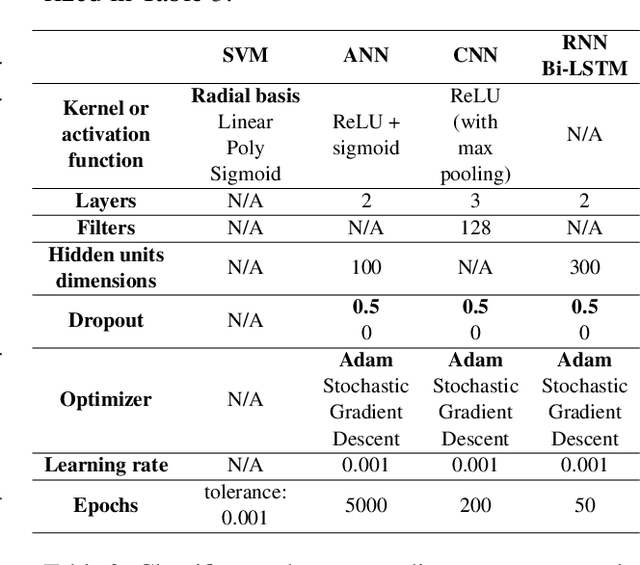
Abstract:Text classification tasks which aim at harvesting and/or organizing information from electronic health records are pivotal to support clinical and translational research. However these present specific challenges compared to other classification tasks, notably due to the particular nature of the medical lexicon and language used in clinical records. Recent advances in embedding methods have shown promising results for several clinical tasks, yet there is no exhaustive comparison of such approaches with other commonly used word representations and classification models. In this work, we analyse the impact of various word representations, text pre-processing and classification algorithms on the performance of four different text classification tasks. The results show that traditional approaches, when tailored to the specific language and structure of the text inherent to the classification task, can achieve or exceed the performance of more recent ones based on contextual embeddings such as BERT.
Identifying physical health comorbidities in a cohort of individuals with severe mental illness: An application of SemEHR
Feb 07, 2020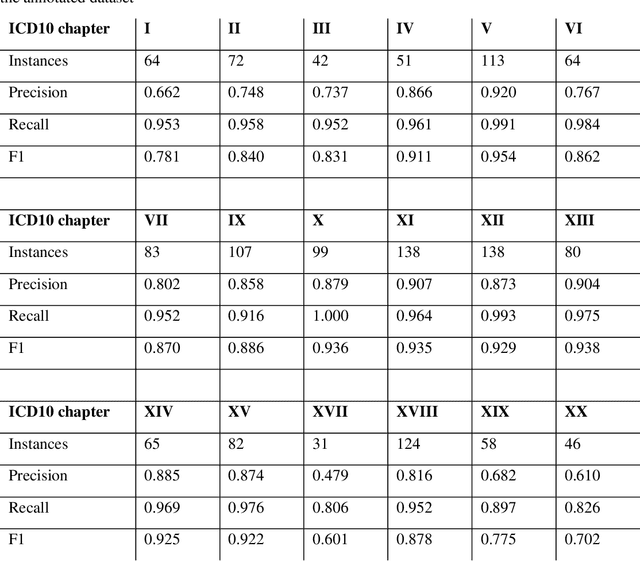
Abstract:Multimorbidity research in mental health services requires data from physical health conditions which is traditionally limited in mental health care electronic health records. In this study, we aimed to extract data from physical health conditions from clinical notes using SemEHR. Data was extracted from Clinical Record Interactive Search (CRIS) system at South London and Maudsley Biomedical Research Centre (SLaM BRC) and the cohort consisted of all individuals who had received a primary or secondary diagnosis of severe mental illness between 2007 and 2018. Three pairs of annotators annotated 2403 documents with an average Cohen's Kappa of 0.757. Results show that the NLP performance varies across different diseases areas (F1 0.601 - 0.954) suggesting that the language patterns or terminologies of different condition groups entail different technical challenges to the same NLP task.
MedCAT -- Medical Concept Annotation Tool
Dec 18, 2019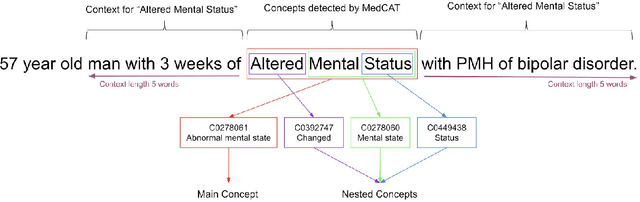
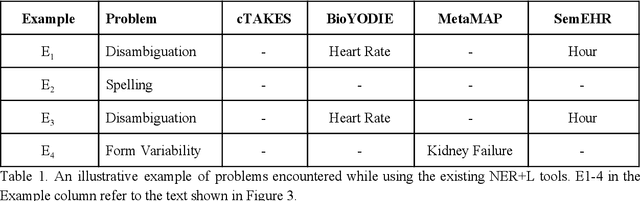
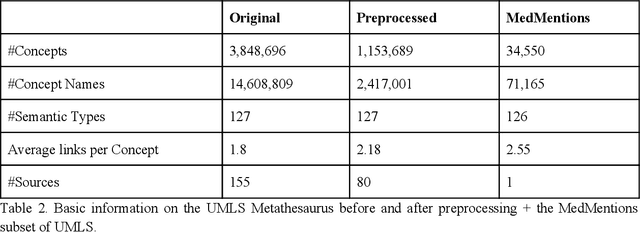
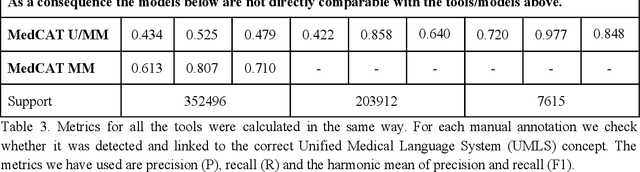
Abstract:Biomedical documents such as Electronic Health Records (EHRs) contain a large amount of information in an unstructured format. The data in EHRs is a hugely valuable resource documenting clinical narratives and decisions, but whilst the text can be easily understood by human doctors it is challenging to use in research and clinical applications. To uncover the potential of biomedical documents we need to extract and structure the information they contain. The task at hand is Named Entity Recognition and Linking (NER+L). The number of entities, ambiguity of words, overlapping and nesting make the biomedical area significantly more difficult than many others. To overcome these difficulties, we have developed the Medical Concept Annotation Tool (MedCAT), an open-source unsupervised approach to NER+L. MedCAT uses unsupervised machine learning to disambiguate entities. It was validated on MIMIC-III (a freely accessible critical care database) and MedMentions (Biomedical papers annotated with mentions from the Unified Medical Language System). In case of NER+L, the comparison with existing tools shows that MedCAT improves the previous best with only unsupervised learning (F1=0.848 vs 0.691 for disease detection; F1=0.710 vs. 0.222 for general concept detection). A qualitative analysis of the vector embeddings learnt by MedCAT shows that it captures latent medical knowledge available in EHRs (MIMIC-III). Unsupervised learning can improve the performance of large scale entity extraction, but it has some limitations when working with only a couple of entities and a small dataset. In that case options are supervised learning or active learning, both of which are supported in MedCAT via the MedCATtrainer extension. Our approach can detect and link millions of different biomedical concepts with state-of-the-art performance, whilst being lightweight, fast and easy to use.
MedCATTrainer: A Biomedical Free Text Annotation Interface with Active Learning and Research Use Case Specific Customisation
Jul 16, 2019
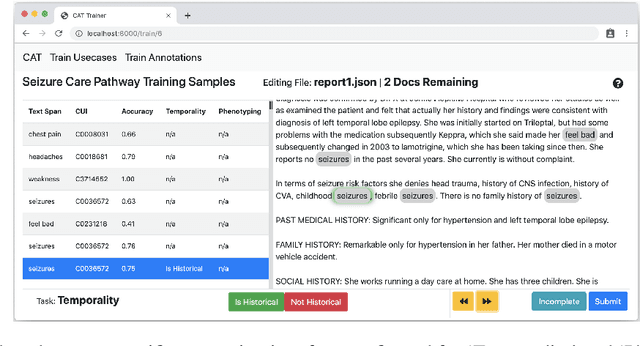
Abstract:We present MedCATTrainer an interface for building, improving and customising a given Named Entity Recognition and Linking (NER+L) model for biomedical domain text. NER+L is often used as a first step in deriving value from clinical text. Collecting labelled data for training models is difficult due to the need for specialist domain knowledge. MedCATTrainer offers an interactive web-interface to inspect and improve recognised entities from an underlying NER+L model via active learning. Secondary use of data for clinical research often has task and context specific criteria. MedCATTrainer provides a further interface to define and collect supervised learning training data for researcher specific use cases. Initial results suggest our approach allows for efficient and accurate collection of research use case specific training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge