Lukasz Roguski
Challenges and Opportunities of Using Transformer-Based Multi-Task Learning in NLP Through ML Lifecycle: A Survey
Aug 16, 2023



Abstract:The increasing adoption of natural language processing (NLP) models across industries has led to practitioners' need for machine learning systems to handle these models efficiently, from training to serving them in production. However, training, deploying, and updating multiple models can be complex, costly, and time-consuming, mainly when using transformer-based pre-trained language models. Multi-Task Learning (MTL) has emerged as a promising approach to improve efficiency and performance through joint training, rather than training separate models. Motivated by this, we first provide an overview of transformer-based MTL approaches in NLP. Then, we discuss the challenges and opportunities of using MTL approaches throughout typical ML lifecycle phases, specifically focusing on the challenges related to data engineering, model development, deployment, and monitoring phases. This survey focuses on transformer-based MTL architectures and, to the best of our knowledge, is novel in that it systematically analyses how transformer-based MTL in NLP fits into ML lifecycle phases. Furthermore, we motivate research on the connection between MTL and continual learning (CL), as this area remains unexplored. We believe it would be practical to have a model that can handle both MTL and CL, as this would make it easier to periodically re-train the model, update it due to distribution shifts, and add new capabilities to meet real-world requirements.
Deployment of a Free-Text Analytics Platform at a UK National Health Service Research Hospital: CogStack at University College London Hospitals
Aug 15, 2021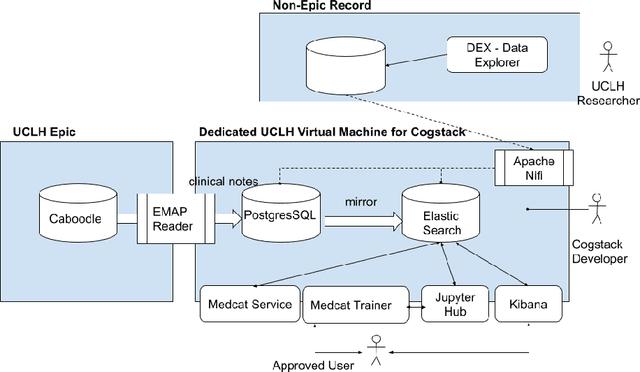
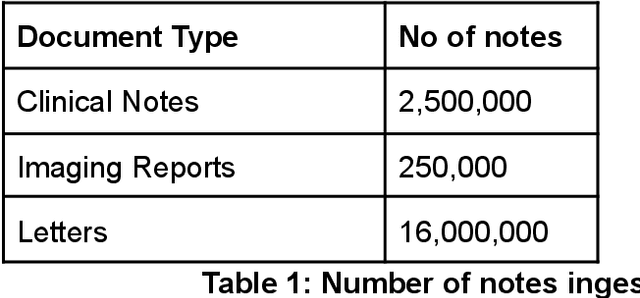

Abstract:As more healthcare organisations transition to using electronic health record (EHR) systems it is important for these organisations to maximise the secondary use of their data to support service improvement and clinical research. These organisations will find it challenging to have systems which can mine information from the unstructured data fields in the record (clinical notes, letters etc) and more practically have such systems interact with all of the hospitals data systems (legacy and current). To tackle this problem at University College London Hospitals, we have deployed an enhanced version of the CogStack platform; an information retrieval platform with natural language processing capabilities which we have configured to process the hospital's existing and legacy records. The platform has improved data ingestion capabilities as well as better tools for natural language processing. To date we have processed over 18 million records and the insights produced from CogStack have informed a number of clinical research use cases at the hospitals.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020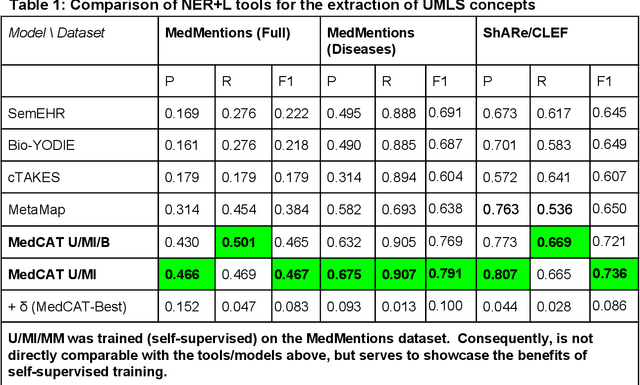
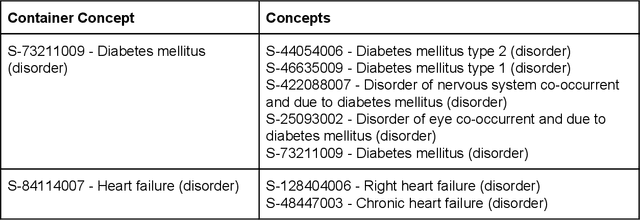
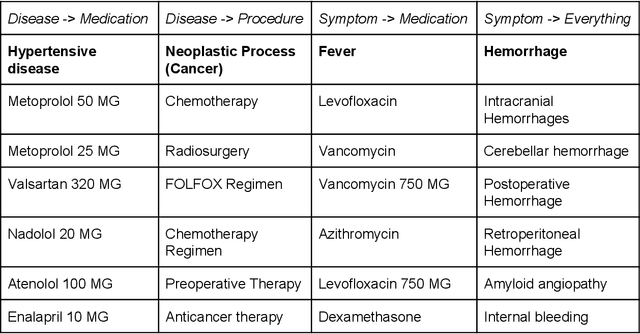
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
MedCAT -- Medical Concept Annotation Tool
Dec 18, 2019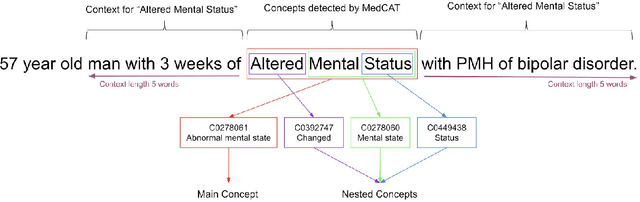
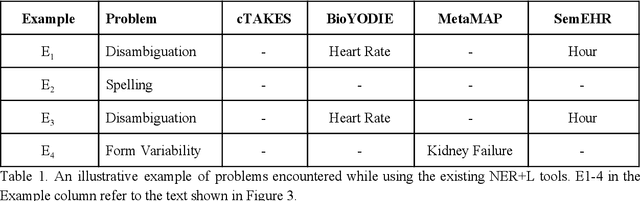
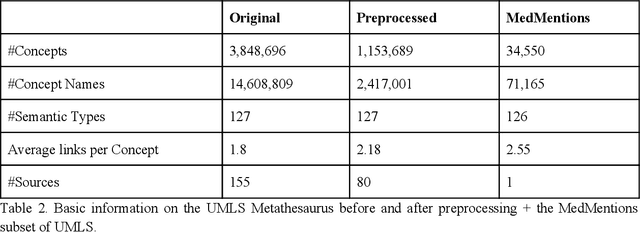
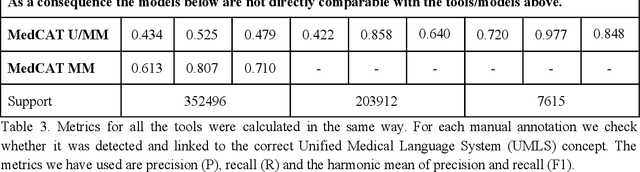
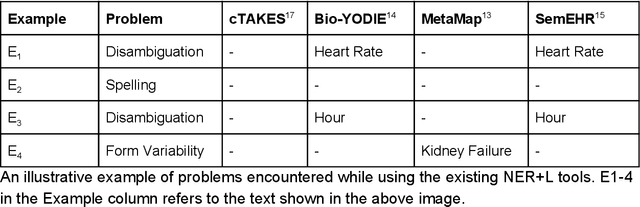
Abstract:Biomedical documents such as Electronic Health Records (EHRs) contain a large amount of information in an unstructured format. The data in EHRs is a hugely valuable resource documenting clinical narratives and decisions, but whilst the text can be easily understood by human doctors it is challenging to use in research and clinical applications. To uncover the potential of biomedical documents we need to extract and structure the information they contain. The task at hand is Named Entity Recognition and Linking (NER+L). The number of entities, ambiguity of words, overlapping and nesting make the biomedical area significantly more difficult than many others. To overcome these difficulties, we have developed the Medical Concept Annotation Tool (MedCAT), an open-source unsupervised approach to NER+L. MedCAT uses unsupervised machine learning to disambiguate entities. It was validated on MIMIC-III (a freely accessible critical care database) and MedMentions (Biomedical papers annotated with mentions from the Unified Medical Language System). In case of NER+L, the comparison with existing tools shows that MedCAT improves the previous best with only unsupervised learning (F1=0.848 vs 0.691 for disease detection; F1=0.710 vs. 0.222 for general concept detection). A qualitative analysis of the vector embeddings learnt by MedCAT shows that it captures latent medical knowledge available in EHRs (MIMIC-III). Unsupervised learning can improve the performance of large scale entity extraction, but it has some limitations when working with only a couple of entities and a small dataset. In that case options are supervised learning or active learning, both of which are supported in MedCAT via the MedCATtrainer extension. Our approach can detect and link millions of different biomedical concepts with state-of-the-art performance, whilst being lightweight, fast and easy to use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge