Wai Keong Wong
Many happy returns: machine learning to support platelet issuing and waste reduction in hospital blood banks
Nov 22, 2024



Abstract:Efforts to reduce platelet wastage in hospital blood banks have focused on ordering policies, but the predominant practice of issuing the oldest unit first may not be optimal when some units are returned unused. We propose a novel, machine learning (ML)-guided issuing policy to increase the likelihood of returned units being reissued before expiration. Our ML model trained to predict returns on 17,297 requests for platelets gave AUROC 0.74 on 9,353 held-out requests. Prior to ML model development we built a simulation of the blood bank operation that incorporated returns to understand the scale of benefits of such a model. Using our trained model in the simulation gave an estimated reduction in wastage of 14%. Our partner hospital is considering adopting our approach, which would be particularly beneficial for hospitals with higher return rates and where units have a shorter remaining useful life on arrival.
Going faster to see further: GPU-accelerated value iteration and simulation for perishable inventory control using JAX
Mar 19, 2023



Abstract:Value iteration can find the optimal replenishment policy for a perishable inventory problem, but is computationally demanding due to the large state spaces that are required to represent the age profile of stock. The parallel processing capabilities of modern GPUs can reduce the wall time required to run value iteration by updating many states simultaneously. The adoption of GPU-accelerated approaches has been limited in operational research relative to other fields like machine learning, in which new software frameworks have made GPU programming widely accessible. We used the Python library JAX to implement value iteration and simulators of the underlying Markov decision processes in a high-level API, and relied on this library's function transformations and compiler to efficiently utilize GPU hardware. Our method can extend use of value iteration to settings that were previously considered infeasible or impractical. We demonstrate this on example scenarios from three recent studies which include problems with over 16 million states and additional problem features, such as substitution between products, that increase computational complexity. We compare the performance of the optimal replenishment policies to heuristic policies, fitted using simulation optimization in JAX which allowed the parallel evaluation of multiple candidate policy parameters on thousands of simulated years. The heuristic policies gave a maximum optimality gap of 2.49%. Our general approach may be applicable to a wide range of problems in operational research that would benefit from large-scale parallel computation on consumer-grade GPU hardware.
Deployment of a Free-Text Analytics Platform at a UK National Health Service Research Hospital: CogStack at University College London Hospitals
Aug 15, 2021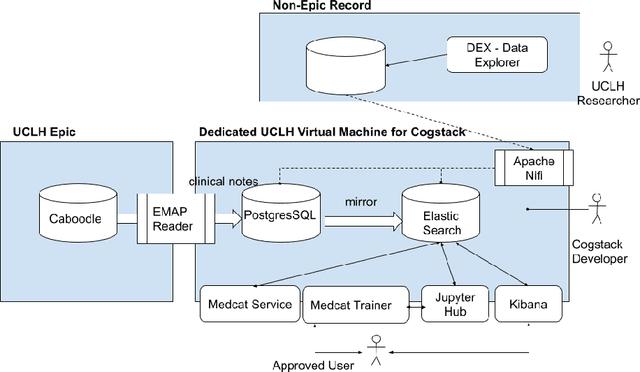
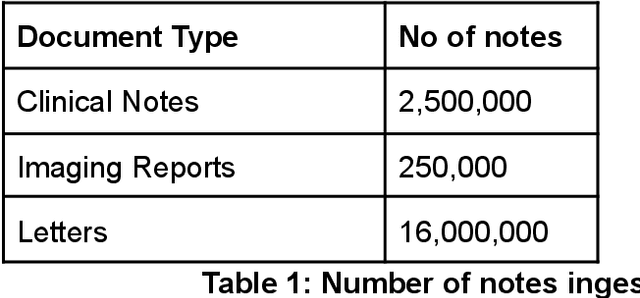

Abstract:As more healthcare organisations transition to using electronic health record (EHR) systems it is important for these organisations to maximise the secondary use of their data to support service improvement and clinical research. These organisations will find it challenging to have systems which can mine information from the unstructured data fields in the record (clinical notes, letters etc) and more practically have such systems interact with all of the hospitals data systems (legacy and current). To tackle this problem at University College London Hospitals, we have deployed an enhanced version of the CogStack platform; an information retrieval platform with natural language processing capabilities which we have configured to process the hospital's existing and legacy records. The platform has improved data ingestion capabilities as well as better tools for natural language processing. To date we have processed over 18 million records and the insights produced from CogStack have informed a number of clinical research use cases at the hospitals.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020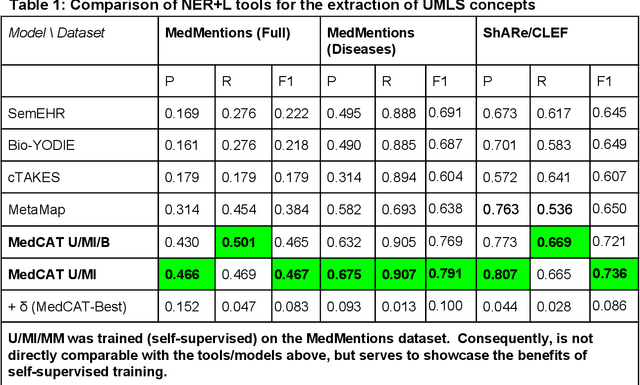
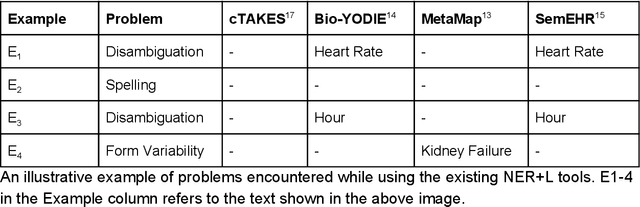
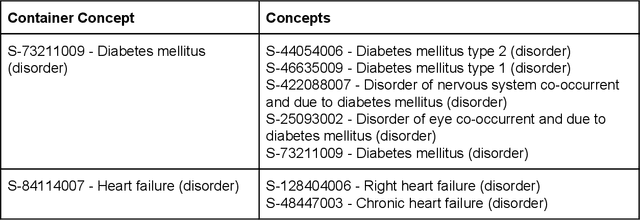
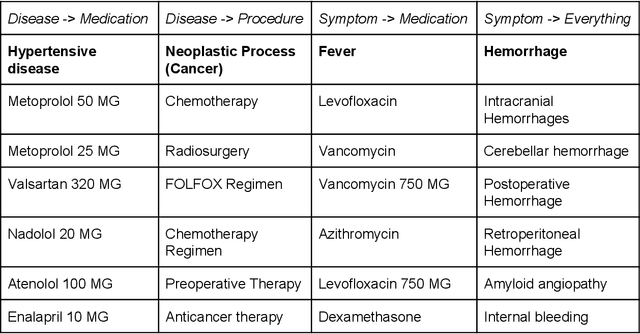
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge