Amos Folarin
Deployment of a Free-Text Analytics Platform at a UK National Health Service Research Hospital: CogStack at University College London Hospitals
Aug 15, 2021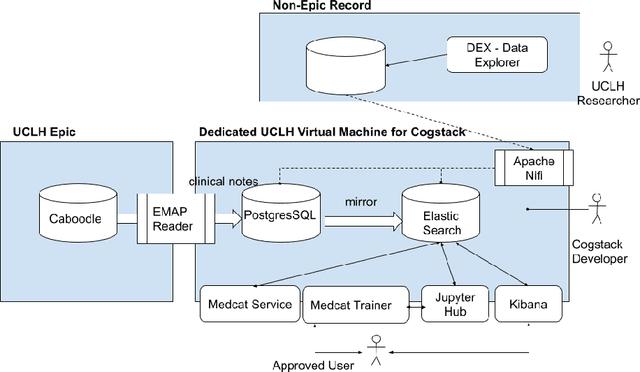
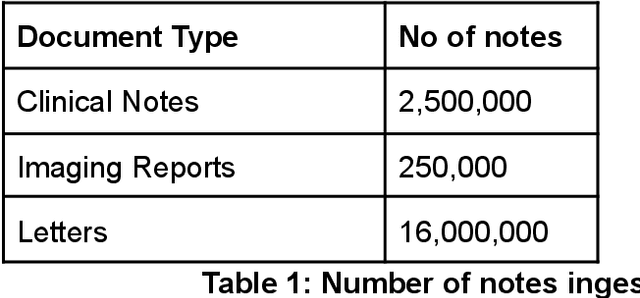

Abstract:As more healthcare organisations transition to using electronic health record (EHR) systems it is important for these organisations to maximise the secondary use of their data to support service improvement and clinical research. These organisations will find it challenging to have systems which can mine information from the unstructured data fields in the record (clinical notes, letters etc) and more practically have such systems interact with all of the hospitals data systems (legacy and current). To tackle this problem at University College London Hospitals, we have deployed an enhanced version of the CogStack platform; an information retrieval platform with natural language processing capabilities which we have configured to process the hospital's existing and legacy records. The platform has improved data ingestion capabilities as well as better tools for natural language processing. To date we have processed over 18 million records and the insights produced from CogStack have informed a number of clinical research use cases at the hospitals.
MedCAT -- Medical Concept Annotation Tool
Dec 18, 2019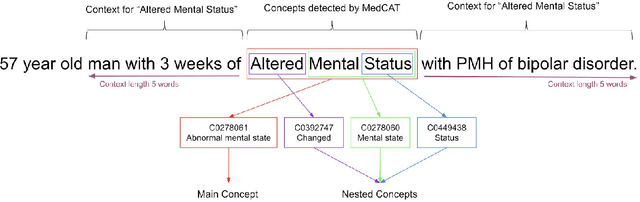
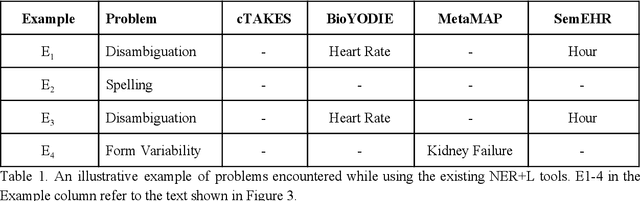
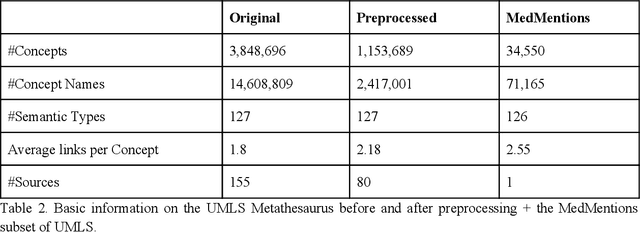
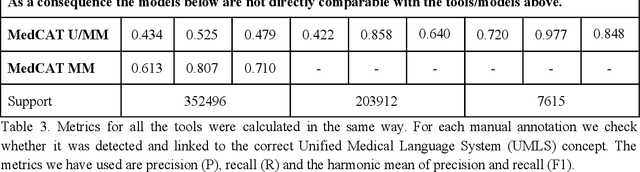
Abstract:Biomedical documents such as Electronic Health Records (EHRs) contain a large amount of information in an unstructured format. The data in EHRs is a hugely valuable resource documenting clinical narratives and decisions, but whilst the text can be easily understood by human doctors it is challenging to use in research and clinical applications. To uncover the potential of biomedical documents we need to extract and structure the information they contain. The task at hand is Named Entity Recognition and Linking (NER+L). The number of entities, ambiguity of words, overlapping and nesting make the biomedical area significantly more difficult than many others. To overcome these difficulties, we have developed the Medical Concept Annotation Tool (MedCAT), an open-source unsupervised approach to NER+L. MedCAT uses unsupervised machine learning to disambiguate entities. It was validated on MIMIC-III (a freely accessible critical care database) and MedMentions (Biomedical papers annotated with mentions from the Unified Medical Language System). In case of NER+L, the comparison with existing tools shows that MedCAT improves the previous best with only unsupervised learning (F1=0.848 vs 0.691 for disease detection; F1=0.710 vs. 0.222 for general concept detection). A qualitative analysis of the vector embeddings learnt by MedCAT shows that it captures latent medical knowledge available in EHRs (MIMIC-III). Unsupervised learning can improve the performance of large scale entity extraction, but it has some limitations when working with only a couple of entities and a small dataset. In that case options are supervised learning or active learning, both of which are supported in MedCAT via the MedCATtrainer extension. Our approach can detect and link millions of different biomedical concepts with state-of-the-art performance, whilst being lightweight, fast and easy to use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge