Amos A Folarin
Large-scale digital phenotyping: identifying depression and anxiety indicators in a general UK population with over 10,000 participants
Sep 24, 2024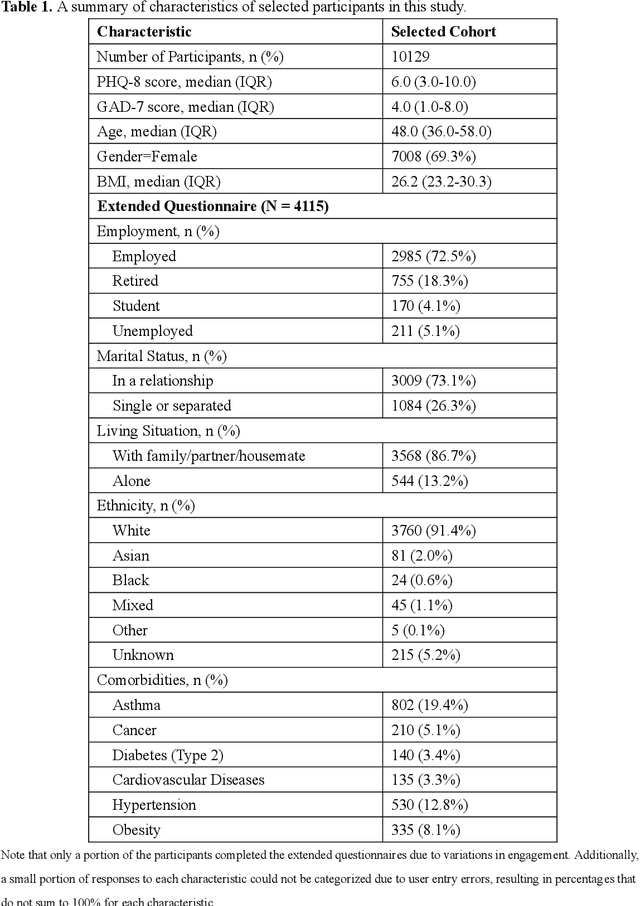



Abstract:Digital phenotyping offers a novel and cost-efficient approach for managing depression and anxiety. Previous studies, often limited to small-to-medium or specific populations, may lack generalizability. We conducted a cross-sectional analysis of data from 10,129 participants recruited from a UK-based general population between June 2020 and August 2022. Participants shared wearable (Fitbit) data and self-reported questionnaires on depression (PHQ-8), anxiety (GAD-7), and mood via a study app. We first examined the correlations between PHQ-8/GAD-7 scores and wearable-derived features, demographics, health data, and mood assessments. Subsequently, unsupervised clustering was used to identify behavioural patterns associated with depression or anxiety. Finally, we employed separate XGBoost models to predict depression and anxiety and compared the results using different subsets of features. We observed significant associations between the severity of depression and anxiety with several factors, including mood, age, gender, BMI, sleep patterns, physical activity, and heart rate. Clustering analysis revealed that participants simultaneously exhibiting lower physical activity levels and higher heart rates reported more severe symptoms. Prediction models incorporating all types of variables achieved the best performance ($R^2$=0.41, MAE=3.42 for depression; $R^2$=0.31, MAE=3.50 for anxiety) compared to those using subsets of variables. This study identified potential indicators for depression and anxiety, highlighting the utility of digital phenotyping and machine learning technologies for rapid screening of mental disorders in general populations. These findings provide robust real-world insights for future healthcare applications.
Identifying depression-related topics in smartphone-collected free-response speech recordings using an automatic speech recognition system and a deep learning topic model
Sep 05, 2023



Abstract:Language use has been shown to correlate with depression, but large-scale validation is needed. Traditional methods like clinic studies are expensive. So, natural language processing has been employed on social media to predict depression, but limitations remain-lack of validated labels, biased user samples, and no context. Our study identified 29 topics in 3919 smartphone-collected speech recordings from 265 participants using the Whisper tool and BERTopic model. Six topics with a median PHQ-8 greater than or equal to 10 were regarded as risk topics for depression: No Expectations, Sleep, Mental Therapy, Haircut, Studying, and Coursework. To elucidate the topic emergence and associations with depression, we compared behavioral (from wearables) and linguistic characteristics across identified topics. The correlation between topic shifts and changes in depression severity over time was also investigated, indicating the importance of longitudinally monitoring language use. We also tested the BERTopic model on a similar smaller dataset (356 speech recordings from 57 participants), obtaining some consistent results. In summary, our findings demonstrate specific speech topics may indicate depression severity. The presented data-driven workflow provides a practical approach to collecting and analyzing large-scale speech data from real-world settings for digital health research.
Disease Insight through Digital Biomarkers Developed by Remotely Collected Wearables and Smartphone Data
Aug 03, 2023



Abstract:Digital Biomarkers and remote patient monitoring can provide valuable and timely insights into how a patient is coping with their condition (disease progression, treatment response, etc.), complementing treatment in traditional healthcare settings.Smartphones with embedded and connected sensors have immense potential for improving healthcare through various apps and mHealth (mobile health) platforms. This capability could enable the development of reliable digital biomarkers from long-term longitudinal data collected remotely from patients. We built an open-source platform, RADAR-base, to support large-scale data collection in remote monitoring studies. RADAR-base is a modern remote data collection platform built around Confluent's Apache Kafka, to support scalability, extensibility, security, privacy and quality of data. It provides support for study design and set-up, active (eg PROMs) and passive (eg. phone sensors, wearable devices and IoT) remote data collection capabilities with feature generation (eg. behavioural, environmental and physiological markers). The backend enables secure data transmission, and scalable solutions for data storage, management and data access. The platform has successfully collected longitudinal data for various cohorts in a number of disease areas including Multiple Sclerosis, Depression, Epilepsy, ADHD, Alzheimer, Autism and Lung diseases. Digital biomarkers developed through collected data are providing useful insights into different diseases. RADAR-base provides a modern open-source, community-driven solution for remote monitoring, data collection, and digital phenotyping of physical and mental health diseases. Clinicians can use digital biomarkers to augment their decision making for the prevention, personalisation and early intervention of disease.
Predicting Depressive Symptom Severity through Individuals' Nearby Bluetooth Devices Count Data Collected by Mobile Phones: A Preliminary Longitudinal Study
Apr 26, 2021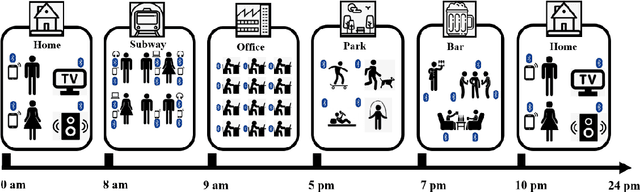
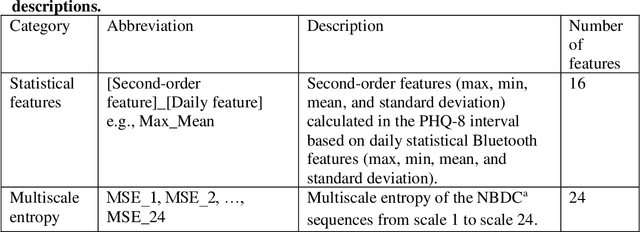
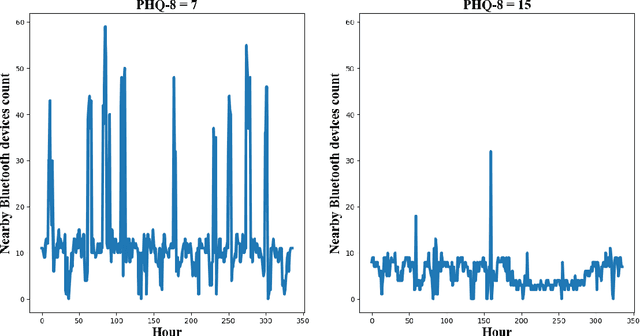
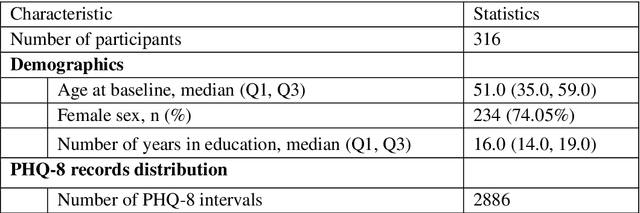
Abstract:The Bluetooth sensor embedded in mobile phones provides an unobtrusive, continuous, and cost-efficient means to capture individuals' proximity information, such as the nearby Bluetooth devices count (NBDC). The continuous NBDC data can partially reflect individuals' behaviors and status, such as social connections and interactions, working status, mobility, and social isolation and loneliness, which were found to be significantly associated with depression by previous survey-based studies. This paper aims to explore the NBDC data's value in predicting depressive symptom severity as measured via the 8-item Patient Health Questionnaire (PHQ-8). The data used in this paper included 2,886 bi-weekly PHQ-8 records collected from 316 participants recruited from three study sites in the Netherlands, Spain, and the UK as part of the EU RADAR-CNS study. From the NBDC data two weeks prior to each PHQ-8 score, we extracted 49 Bluetooth features, including statistical features and nonlinear features for measuring periodicity and regularity of individuals' life rhythms. Linear mixed-effect models were used to explore associations between Bluetooth features and the PHQ-8 score. We then applied hierarchical Bayesian linear regression models to predict the PHQ-8 score from the extracted Bluetooth features. A number of significant associations were found between Bluetooth features and depressive symptom severity. Compared with commonly used machine learning models, the proposed hierarchical Bayesian linear regression model achieved the best prediction metrics, R2= 0.526, and root mean squared error (RMSE) of 3.891. Bluetooth features can explain an extra 18.8% of the variance in the PHQ-8 score relative to the baseline model without Bluetooth features (R2=0.338, RMSE = 4.547).
Fitbeat: COVID-19 Estimation based on Wristband Heart Rate
Apr 19, 2021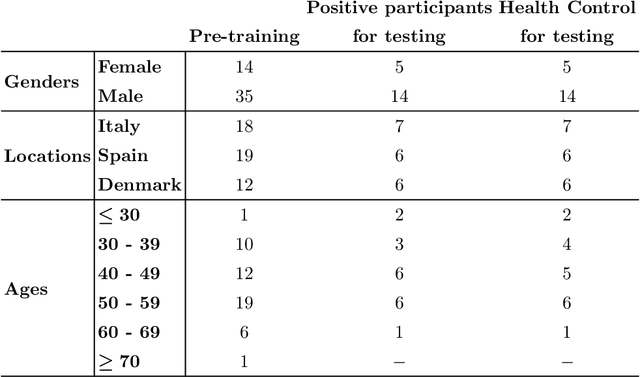

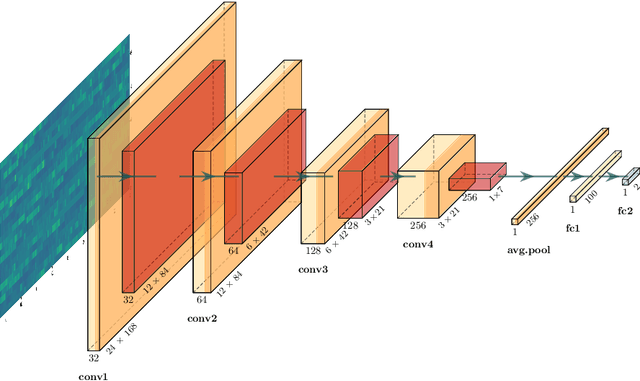
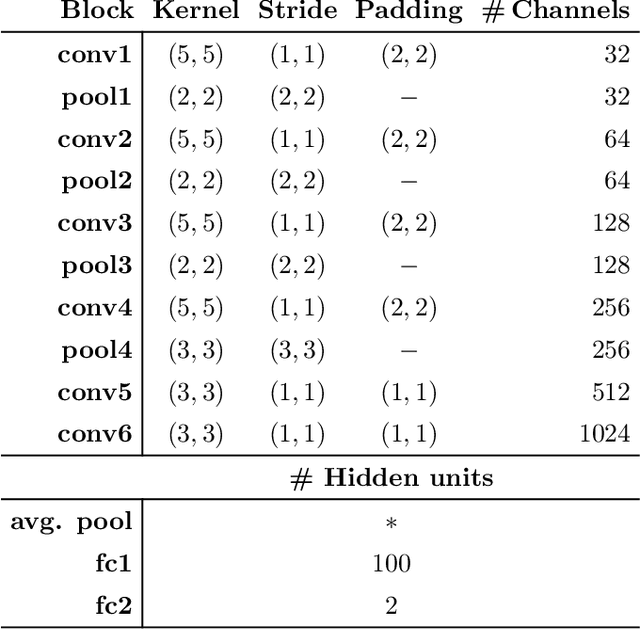
Abstract:This study investigates the potential of deep learning methods to identify individuals with suspected COVID-19 infection using remotely collected heart-rate data. The study utilises data from the ongoing EU IMI RADAR-CNS research project that is investigating the feasibility of wearable devices and smart phones to monitor individuals with multiple sclerosis (MS), depression or epilepsy. Aspart of the project protocol, heart-rate data was collected from participants using a Fitbit wristband. The presence of COVID-19 in the cohort in this work was either confirmed through a positive swab test, or inferred through the self-reporting of a combination of symptoms including fever, respiratory symptoms, loss of smell or taste, tiredness and gastrointestinal symptoms. Experimental results indicate that our proposed contrastive convolutional auto-encoder (contrastive CAE), i. e., a combined architecture of an auto-encoder and contrastive loss, outperforms a conventional convolutional neural network (CNN), as well as a convolutional auto-encoder (CAE) without using contrastive loss. Our final contrastive CAE achieves 95.3% unweighted average recall, 86.4% precision, anF1 measure of 88.2%, a sensitivity of 100% and a specificity of 90.6% on a testset of 19 participants with MS who reported symptoms of COVID-19. Each of these participants was paired with a participant with MS with no COVID-19 symptoms.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020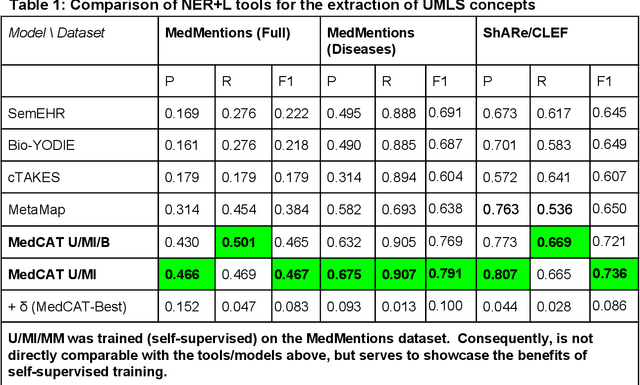
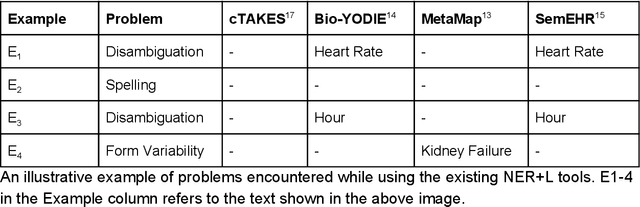
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge