James T Teo
Foresight -- Deep Generative Modelling of Patient Timelines using Electronic Health Records
Dec 13, 2022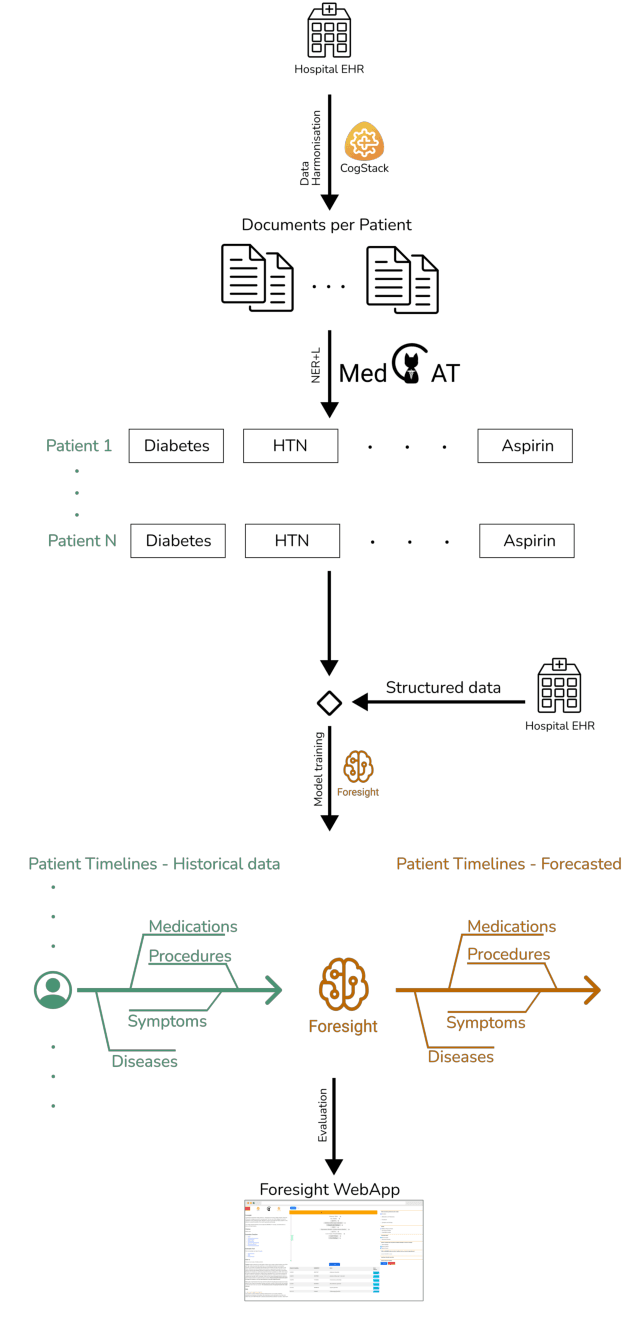
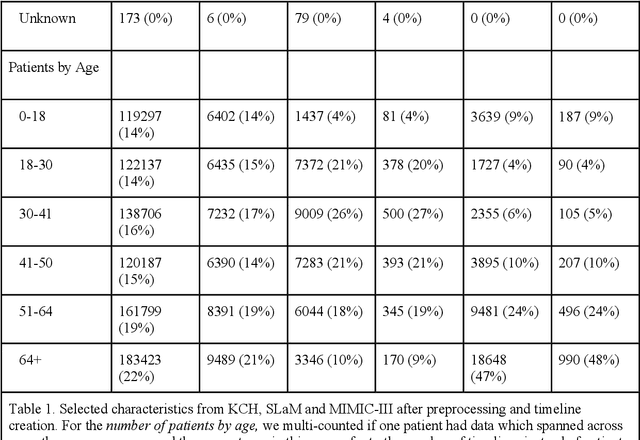
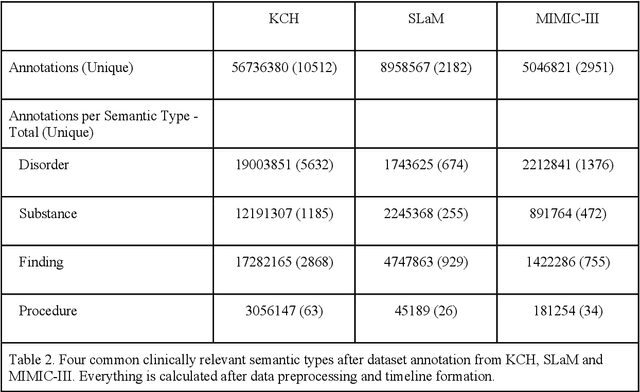
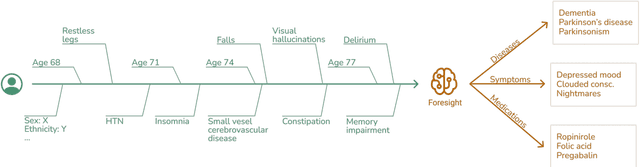
Abstract:Electronic Health Records (EHRs) hold detailed longitudinal information about each patient's health status and general clinical history, a large portion of which is stored within the unstructured text. Temporal modelling of this medical history, which considers the sequence of events, can be used to forecast and simulate future events, estimate risk, suggest alternative diagnoses or forecast complications. While most prediction approaches use mainly structured data or a subset of single-domain forecasts and outcomes, we processed the entire free-text portion of EHRs for longitudinal modelling. We present Foresight, a novel GPT3-based pipeline that uses NER+L tools (i.e. MedCAT) to convert document text into structured, coded concepts, followed by providing probabilistic forecasts for future medical events such as disorders, medications, symptoms and interventions. Since large portions of EHR data are in text form, such an approach benefits from a granular and detailed view of a patient while introducing modest additional noise. On tests in two large UK hospitals (King's College Hospital, South London and Maudsley) and the US MIMIC-III dataset precision@10 of 0.80, 0.81 and 0.91 was achieved for forecasting the next biomedical concept. Foresight was also validated on 34 synthetic patient timelines by 5 clinicians and achieved relevancy of 97% for the top forecasted candidate disorder. Foresight can be easily trained and deployed locally as it only requires free-text data (as a minimum). As a generative model, it can simulate follow-on disorders, medications and interventions for as many steps as required. Foresight is a general-purpose model for biomedical concept modelling that can be used for real-world risk estimation, virtual trials and clinical research to study the progression of diseases, simulate interventions and counterfactuals, and for educational purposes.
A Knowledge Distillation Ensemble Framework for Predicting Short and Long-term Hospitalisation Outcomes from Electronic Health Records Data
Nov 18, 2020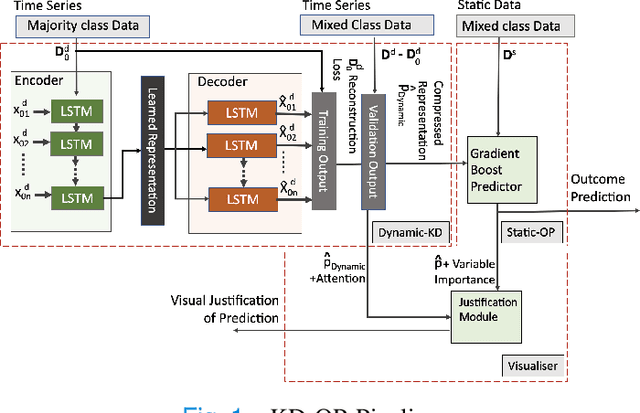
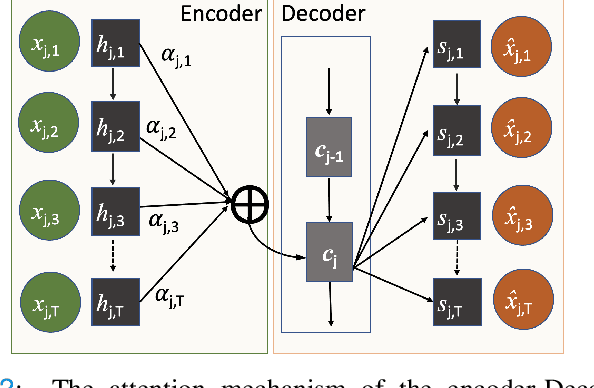
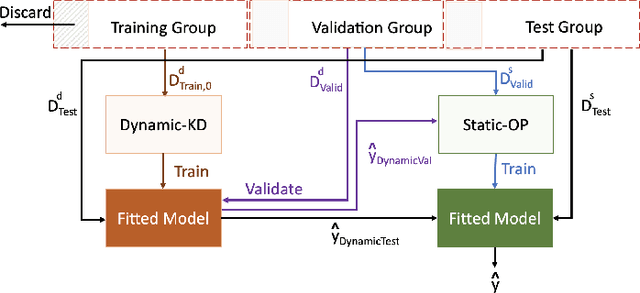
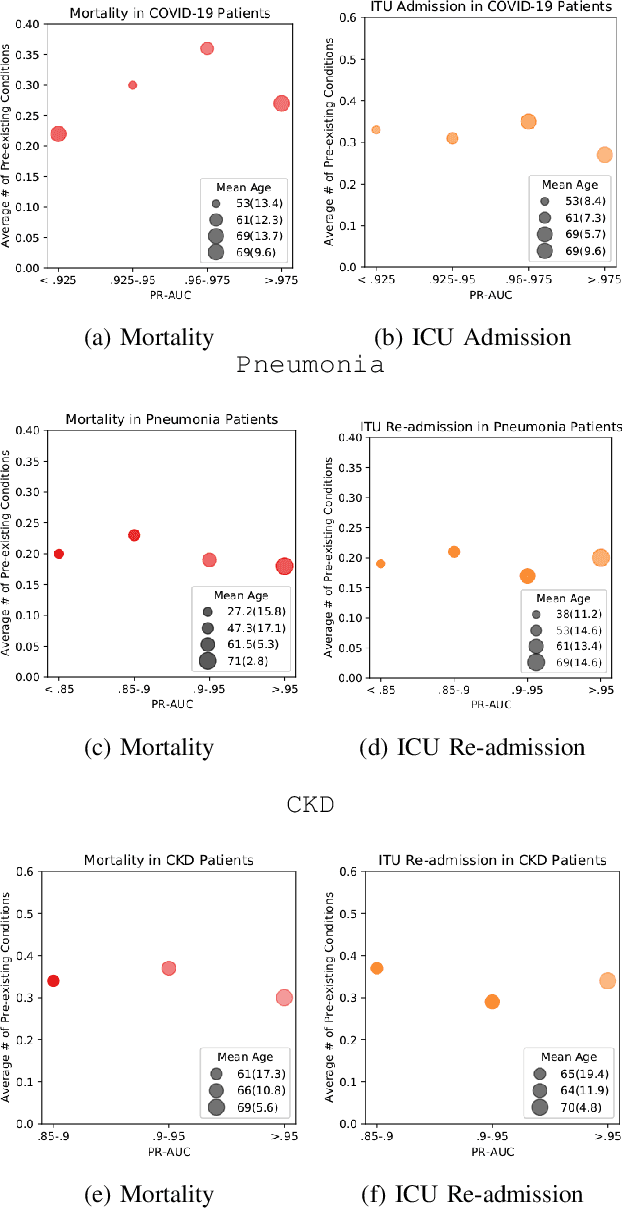
Abstract:The ability to perform accurate prognosis of patients is crucial for proactive clinical decision making, informed resource management and personalised care. Existing outcome prediction models suffer from a low recall of infrequent positive outcomes. We present a highly-scalable and robust machine learning framework to automatically predict adversity represented by mortality and ICU admission from time-series vital signs and laboratory results obtained within the first 24 hours of hospital admission. The stacked platform comprises two components: a) an unsupervised LSTM Autoencoder that learns an optimal representation of the time-series, using it to differentiate the less frequent patterns which conclude with an adverse event from the majority patterns that do not, and b) a gradient boosting model, which relies on the constructed representation to refine prediction, incorporating static features of demographics, admission details and clinical summaries. The model is used to assess a patient's risk of adversity over time and provides visual justifications of its prediction based on the patient's static features and dynamic signals. Results of three case studies for predicting mortality and ICU admission show that the model outperforms all existing outcome prediction models, achieving PR-AUC of 0.93 (95$%$ CI: 0.878 - 0.969) in predicting mortality in ICU and general ward settings and 0.987 (95$%$ CI: 0.985-0.995) in predicting ICU admission.
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020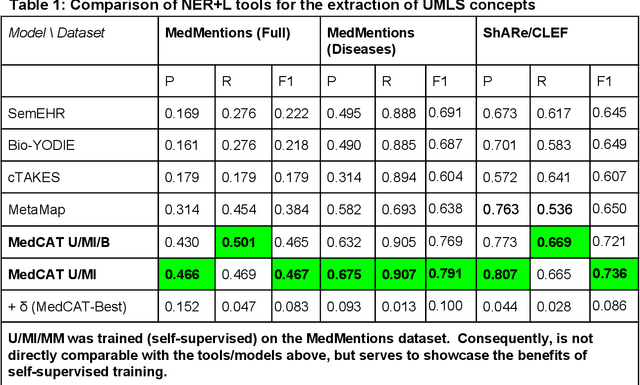
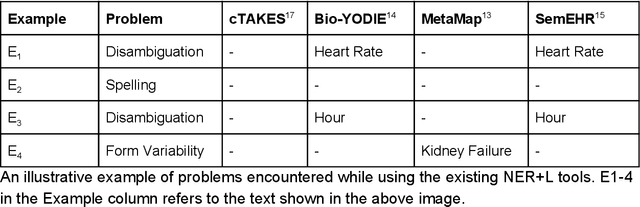
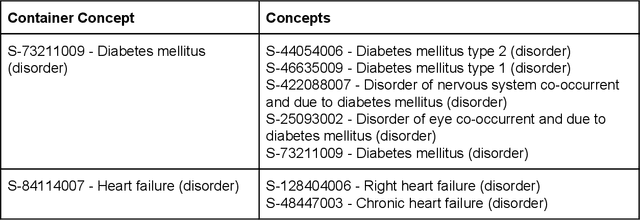
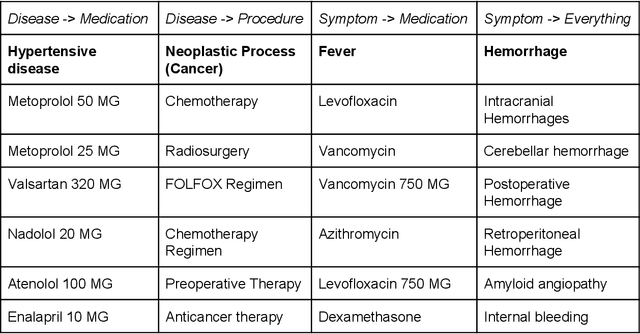
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge