Joshua Au Yeung
Large Language Models for Medical Forecasting -- Foresight 2
Dec 14, 2024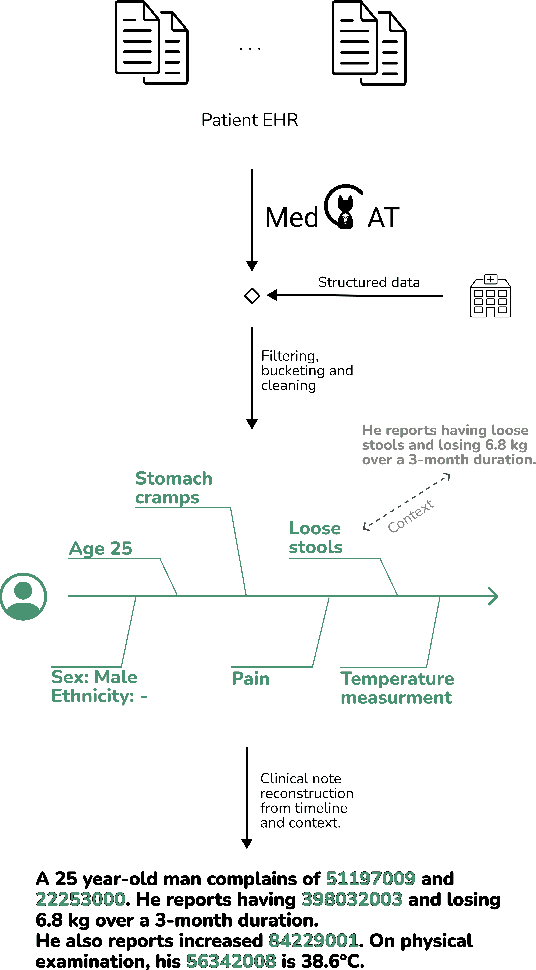
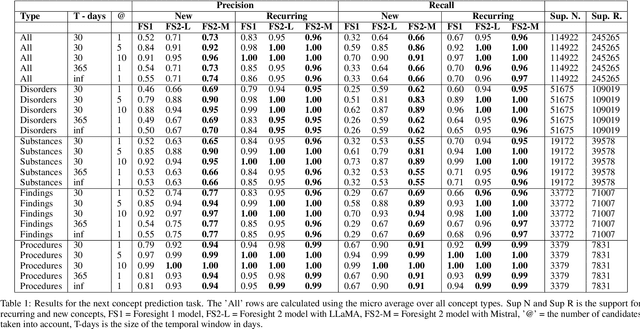
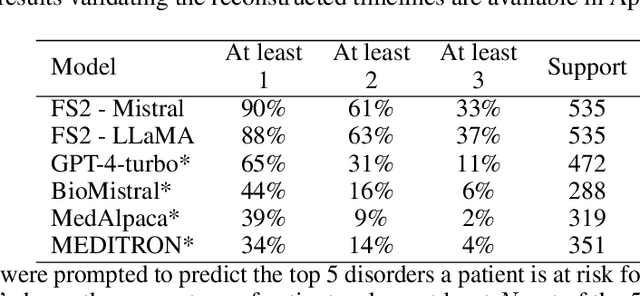
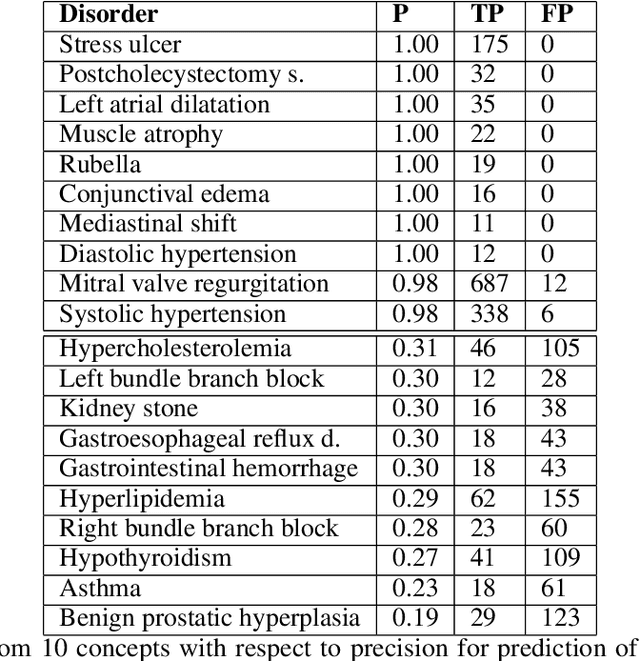
Abstract:Foresight 2 (FS2) is a large language model fine-tuned on hospital data for modelling patient timelines (GitHub 'removed for anon'). It can understand patients' clinical notes and predict SNOMED codes for a wide range of biomedical use cases, including diagnosis suggestions, risk forecasting, and procedure and medication recommendations. FS2 is trained on the free text portion of the MIMIC-III dataset, firstly through extracting biomedical concepts and then creating contextualised patient timelines, upon which the model is then fine-tuned. The results show significant improvement over the previous state-of-the-art for the next new biomedical concept prediction (P/R - 0.73/0.66 vs 0.52/0.32) and a similar improvement specifically for the next new disorder prediction (P/R - 0.69/0.62 vs 0.46/0.25). Finally, on the task of risk forecast, we compare our model to GPT-4-turbo (and a range of open-source biomedical LLMs) and show that FS2 performs significantly better on such tasks (P@5 - 0.90 vs 0.65). This highlights the need to incorporate hospital data into LLMs and shows that small models outperform much larger ones when fine-tuned on high-quality, specialised data.
Validating transformers for redaction of text from electronic health records in real-world healthcare
Oct 05, 2023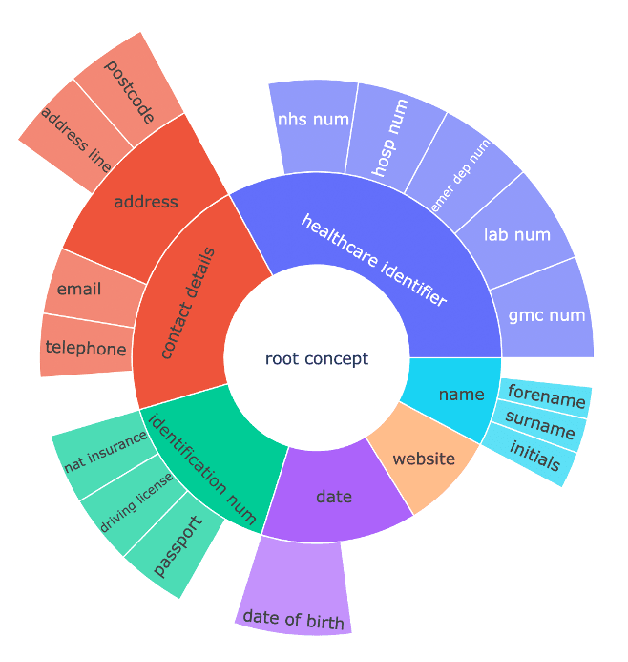
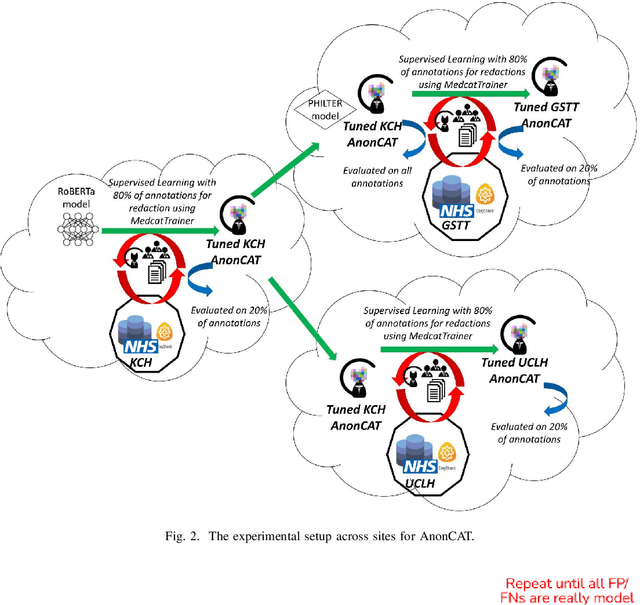
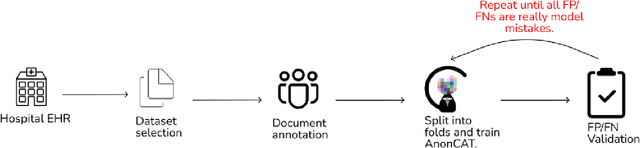
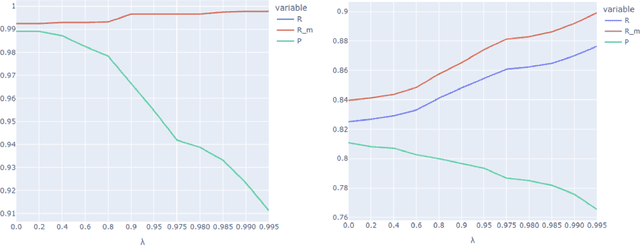
Abstract:Protecting patient privacy in healthcare records is a top priority, and redaction is a commonly used method for obscuring directly identifiable information in text. Rule-based methods have been widely used, but their precision is often low causing over-redaction of text and frequently not being adaptable enough for non-standardised or unconventional structures of personal health information. Deep learning techniques have emerged as a promising solution, but implementing them in real-world environments poses challenges due to the differences in patient record structure and language across different departments, hospitals, and countries. In this study, we present AnonCAT, a transformer-based model and a blueprint on how deidentification models can be deployed in real-world healthcare. AnonCAT was trained through a process involving manually annotated redactions of real-world documents from three UK hospitals with different electronic health record systems and 3116 documents. The model achieved high performance in all three hospitals with a Recall of 0.99, 0.99 and 0.96. Our findings demonstrate the potential of deep learning techniques for improving the efficiency and accuracy of redaction in global healthcare data and highlight the importance of building workflows which not just use these models but are also able to continually fine-tune and audit the performance of these algorithms to ensure continuing effectiveness in real-world settings. This approach provides a blueprint for the real-world use of de-identifying algorithms through fine-tuning and localisation, the code together with tutorials is available on GitHub (https://github.com/CogStack/MedCAT).
Foresight -- Deep Generative Modelling of Patient Timelines using Electronic Health Records
Dec 13, 2022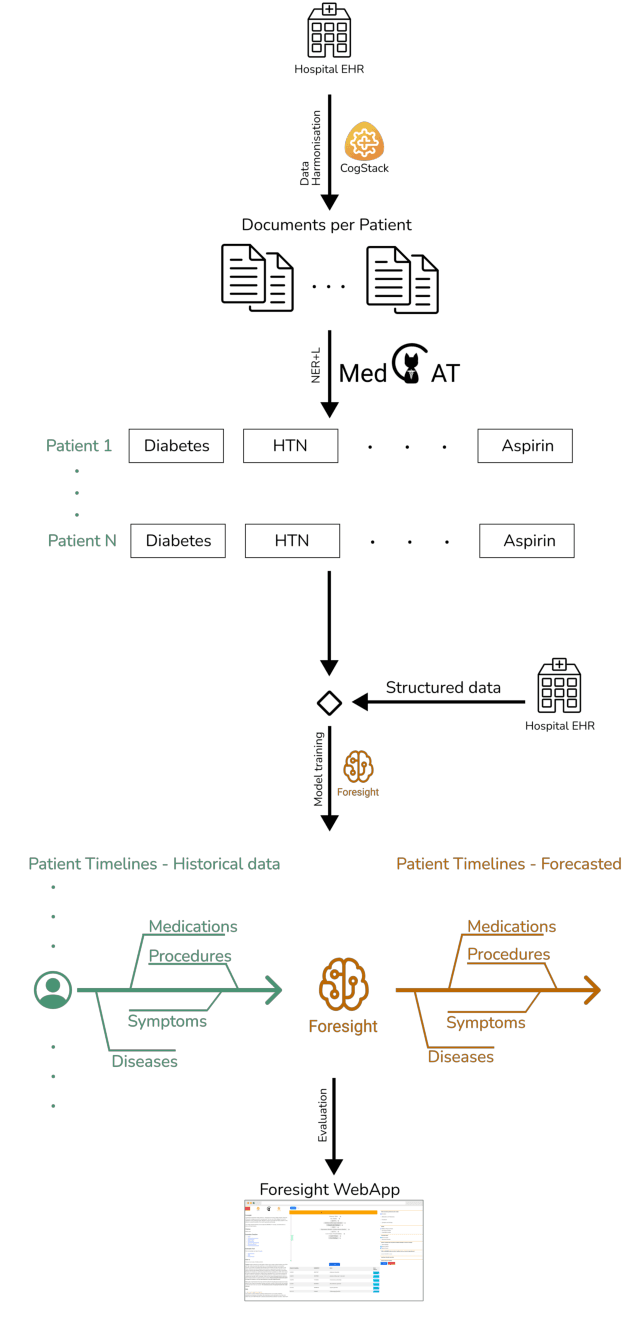
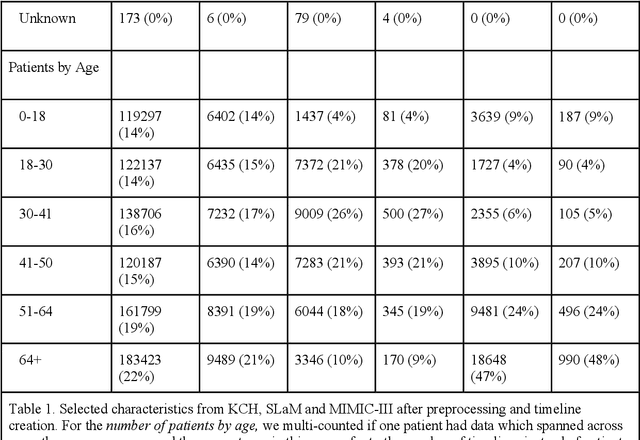
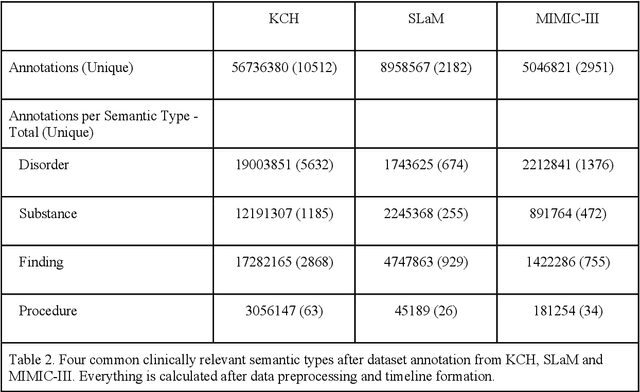
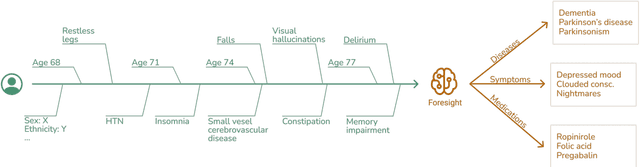
Abstract:Electronic Health Records (EHRs) hold detailed longitudinal information about each patient's health status and general clinical history, a large portion of which is stored within the unstructured text. Temporal modelling of this medical history, which considers the sequence of events, can be used to forecast and simulate future events, estimate risk, suggest alternative diagnoses or forecast complications. While most prediction approaches use mainly structured data or a subset of single-domain forecasts and outcomes, we processed the entire free-text portion of EHRs for longitudinal modelling. We present Foresight, a novel GPT3-based pipeline that uses NER+L tools (i.e. MedCAT) to convert document text into structured, coded concepts, followed by providing probabilistic forecasts for future medical events such as disorders, medications, symptoms and interventions. Since large portions of EHR data are in text form, such an approach benefits from a granular and detailed view of a patient while introducing modest additional noise. On tests in two large UK hospitals (King's College Hospital, South London and Maudsley) and the US MIMIC-III dataset precision@10 of 0.80, 0.81 and 0.91 was achieved for forecasting the next biomedical concept. Foresight was also validated on 34 synthetic patient timelines by 5 clinicians and achieved relevancy of 97% for the top forecasted candidate disorder. Foresight can be easily trained and deployed locally as it only requires free-text data (as a minimum). As a generative model, it can simulate follow-on disorders, medications and interventions for as many steps as required. Foresight is a general-purpose model for biomedical concept modelling that can be used for real-world risk estimation, virtual trials and clinical research to study the progression of diseases, simulate interventions and counterfactuals, and for educational purposes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge