Robert Stewart
Determinants of Training Corpus Size for Clinical Text Classification
Jan 22, 2026Abstract:Introduction: Clinical text classification using natural language processing (NLP) models requires adequate training data to achieve optimal performance. For that, 200-500 documents are typically annotated. The number is constrained by time and costs and lacks justification of the sample size requirements and their relationship to text vocabulary properties. Methods: Using the publicly available MIMIC-III dataset containing hospital discharge notes with ICD-9 diagnoses as labels, we employed pre-trained BERT embeddings followed by Random Forest classifiers to identify 10 randomly selected diagnoses, varying training corpus sizes from 100 to 10,000 documents, and analyzed vocabulary properties by identifying strong and noisy predictive words through Lasso logistic regression on bag-of-words embeddings. Results: Learning curves varied significantly across the 10 classification tasks despite identical preprocessing and algorithms, with 600 documents sufficient to achieve 95% of the performance attainable with 10,000 documents for all tasks. Vocabulary analysis revealed that more strong predictors and fewer noisy predictors were associated with steeper learning curves, where every 100 additional noisy words decreased accuracy by approximately 0.02 while 100 additional strong predictors increased maximum accuracy by approximately 0.04.
Sample Size in Natural Language Processing within Healthcare Research
Sep 05, 2023



Abstract:Sample size calculation is an essential step in most data-based disciplines. Large enough samples ensure representativeness of the population and determine the precision of estimates. This is true for most quantitative studies, including those that employ machine learning methods, such as natural language processing, where free-text is used to generate predictions and classify instances of text. Within the healthcare domain, the lack of sufficient corpora of previously collected data can be a limiting factor when determining sample sizes for new studies. This paper tries to address the issue by making recommendations on sample sizes for text classification tasks in the healthcare domain. Models trained on the MIMIC-III database of critical care records from Beth Israel Deaconess Medical Center were used to classify documents as having or not having Unspecified Essential Hypertension, the most common diagnosis code in the database. Simulations were performed using various classifiers on different sample sizes and class proportions. This was repeated for a comparatively less common diagnosis code within the database of diabetes mellitus without mention of complication. Smaller sample sizes resulted in better results when using a K-nearest neighbours classifier, whereas larger sample sizes provided better results with support vector machines and BERT models. Overall, a sample size larger than 1000 was sufficient to provide decent performance metrics. The simulations conducted within this study provide guidelines that can be used as recommendations for selecting appropriate sample sizes and class proportions, and for predicting expected performance, when building classifiers for textual healthcare data. The methodology used here can be modified for sample size estimates calculations with other datasets.
Development of a Knowledge Graph Embeddings Model for Pain
Aug 17, 2023



Abstract:Pain is a complex concept that can interconnect with other concepts such as a disorder that might cause pain, a medication that might relieve pain, and so on. To fully understand the context of pain experienced by either an individual or across a population, we may need to examine all concepts related to pain and the relationships between them. This is especially useful when modeling pain that has been recorded in electronic health records. Knowledge graphs represent concepts and their relations by an interlinked network, enabling semantic and context-based reasoning in a computationally tractable form. These graphs can, however, be too large for efficient computation. Knowledge graph embeddings help to resolve this by representing the graphs in a low-dimensional vector space. These embeddings can then be used in various downstream tasks such as classification and link prediction. The various relations associated with pain which are required to construct such a knowledge graph can be obtained from external medical knowledge bases such as SNOMED CT, a hierarchical systematic nomenclature of medical terms. A knowledge graph built in this way could be further enriched with real-world examples of pain and its relations extracted from electronic health records. This paper describes the construction of such knowledge graph embedding models of pain concepts, extracted from the unstructured text of mental health electronic health records, combined with external knowledge created from relations described in SNOMED CT, and their evaluation on a subject-object link prediction task. The performance of the models was compared with other baseline models.
Identifying Mentions of Pain in Mental Health Records Text: A Natural Language Processing Approach
Apr 05, 2023

Abstract:Pain is a common reason for accessing healthcare resources and is a growing area of research, especially in its overlap with mental health. Mental health electronic health records are a good data source to study this overlap. However, much information on pain is held in the free text of these records, where mentions of pain present a unique natural language processing problem due to its ambiguous nature. This project uses data from an anonymised mental health electronic health records database. The data are used to train a machine learning based classification algorithm to classify sentences as discussing patient pain or not. This will facilitate the extraction of relevant pain information from large databases, and the use of such outputs for further studies on pain and mental health. 1,985 documents were manually triple-annotated for creation of gold standard training data, which was used to train three commonly used classification algorithms. The best performing model achieved an F1-score of 0.98 (95% CI 0.98-0.99).
Logic of Differentiable Logics: Towards a Uniform Semantics of DL
Mar 19, 2023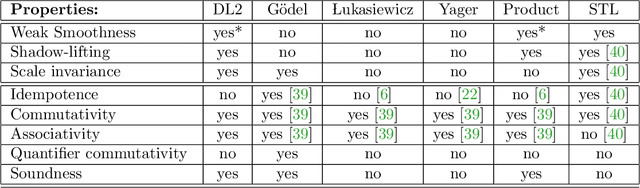
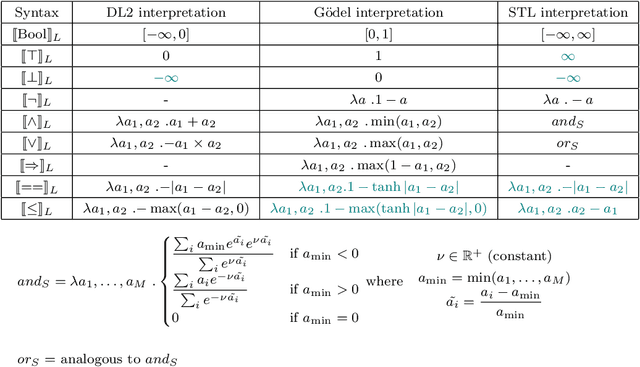
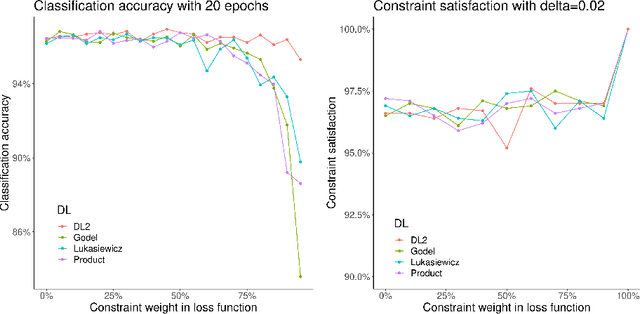
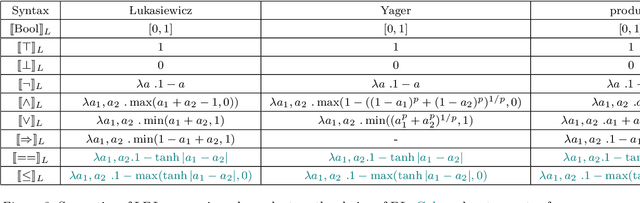
Abstract:Differentiable logics (DL) have recently been proposed as a method of training neural networks to satisfy logical specifications. A DL consists of a syntax in which specifications are stated and an interpretation function that translates expressions in the syntax into loss functions. These loss functions can then be used during training with standard gradient descent algorithms. The variety of existing DLs and the differing levels of formality with which they are treated makes a systematic comparative study of their properties and implementations difficult. This paper remedies this problem by suggesting a meta-language for defining DLs that we call the Logic of Differentiable Logics, or LDL. Syntactically, it generalises the syntax of existing DLs to FOL, and for the first time introduces the formalism for reasoning about vectors and learners. Semantically, it introduces a general interpretation function that can be instantiated to define loss functions arising from different existing DLs. We use LDL to establish several theoretical properties of existing DLs, and to conduct their empirical study in neural network verification.
Differentiable Logics for Neural Network Training and Verification
Jul 14, 2022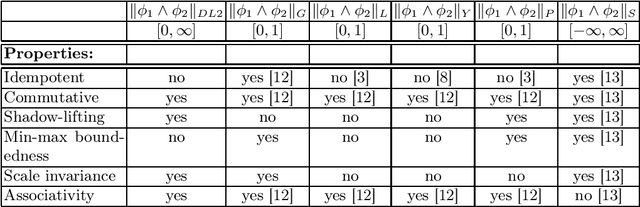
Abstract:The rising popularity of neural networks (NNs) in recent years and their increasing prevalence in real-world applications have drawn attention to the importance of their verification. While verification is known to be computationally difficult theoretically, many techniques have been proposed for solving it in practice. It has been observed in the literature that by default neural networks rarely satisfy logical constraints that we want to verify. A good course of action is to train the given NN to satisfy said constraint prior to verifying them. This idea is sometimes referred to as continuous verification, referring to the loop between training and verification. Usually training with constraints is implemented by specifying a translation for a given formal logic language into loss functions. These loss functions are then used to train neural networks. Because for training purposes these functions need to be differentiable, these translations are called differentiable logics (DL). This raises several research questions. What kind of differentiable logics are possible? What difference does a specific choice of DL make in the context of continuous verification? What are the desirable criteria for a DL viewed from the point of view of the resulting loss function? In this extended abstract we will discuss and answer these questions.
Benchmarking Quantized Neural Networks on FPGAs with FINN
Feb 02, 2021



Abstract:The ever-growing cost of both training and inference for state-of-the-art neural networks has brought literature to look upon ways to cut off resources used with a minimal impact on accuracy. Using lower precision comes at the cost of negligible loss in accuracy. While training neural networks may require a powerful setup, deploying a network must be possible on low-power and low-resource hardware architectures. Reconfigurable architectures have proven to be more powerful and flexible than GPUs when looking at a specific application. This article aims to assess the impact of mixed-precision when applied to neural networks deployed on FPGAs. While several frameworks exist that create tools to deploy neural networks using reduced-precision, few of them assess the importance of quantization and the framework quality. FINN and Brevitas, two frameworks from Xilinx labs, are used to assess the impact of quantization on neural networks using 2 to 8 bit precisions and weights with several parallelization configurations. Equivalent accuracy can be obtained using lower-precision representation and enough training. However, the compressed network can be better parallelized allowing the deployed network throughput to be 62 times faster. The benchmark set up in this work is available in a public repository (https://github.com/QDucasse/nn benchmark).
Multi-domain Clinical Natural Language Processing with MedCAT: the Medical Concept Annotation Toolkit
Oct 02, 2020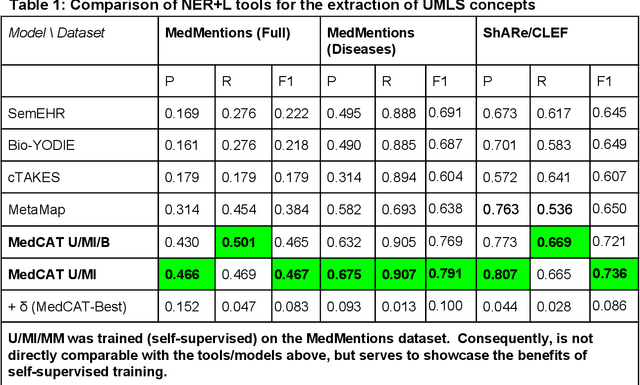
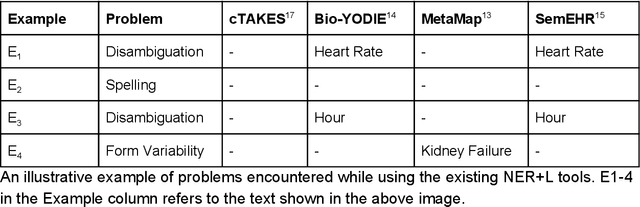
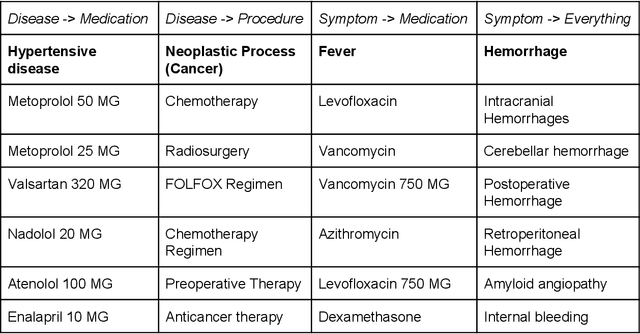
Abstract:Electronic health records (EHR) contain large volumes of unstructured text, requiring the application of Information Extraction (IE) technologies to enable clinical analysis. We present the open source Medical Concept Annotation Toolkit (MedCAT) that provides: a) a novel self-supervised machine learning algorithm for extracting concepts using any concept vocabulary including UMLS/SNOMED-CT; b) a feature-rich annotation interface for customizing and training IE models; and c) integrations to the broader CogStack ecosystem for vendor-agnostic health system deployment. We show improved performance in extracting UMLS concepts from open datasets ( F1 0.467-0.791 vs 0.384-0.691). Further real-world validation demonstrates SNOMED-CT extraction at 3 large London hospitals with self-supervised training over ~8.8B words from ~17M clinical records and further fine-tuning with ~6K clinician annotated examples. We show strong transferability ( F1 >0.94) between hospitals, datasets and concept types indicating cross-domain EHR-agnostic utility for accelerated clinical and research use cases.
Bombus Species Image Classification
Jun 09, 2020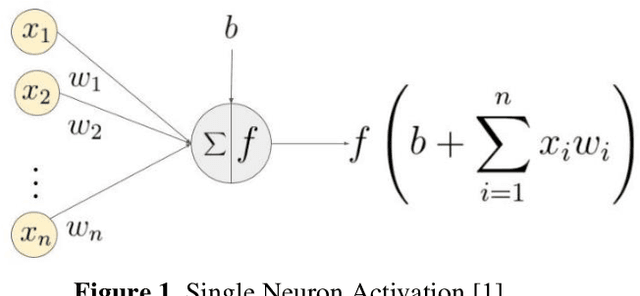
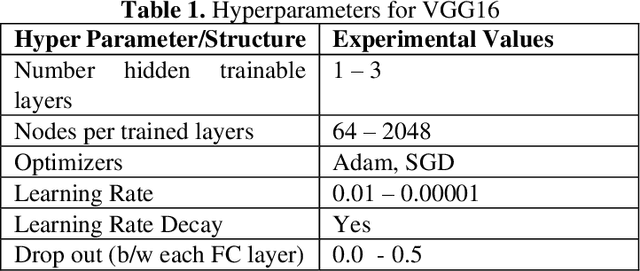
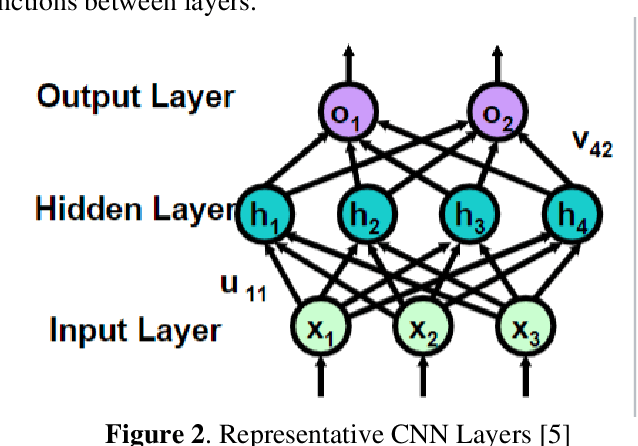
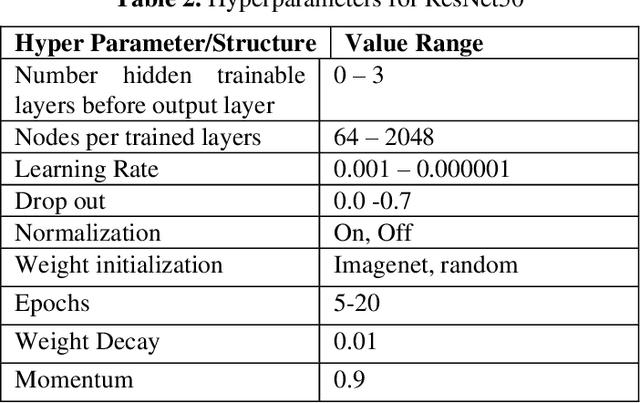
Abstract:Entomologists, ecologists and others struggle to rapidly and accurately identify the species of bumble bees they encounter in their field work and research. The current process requires the bees to be mounted, then physically shipped to a taxonomic expert for proper categorization. We investigated whether an image classification system derived from transfer learning can do this task. We used Google Inception, Oxford VGG16 and VGG19 and Microsoft ResNet 50. We found Inception and VGG classifiers were able to make some progress at identifying bumble bee species from the available data, whereas ResNet was not. Individual classifiers achieved accuracies of up to 23% for single species identification and 44% top-3 labels, where a composite model performed better, 27% and 50%. We feel the performance was most hampered by our limited data set of 5,000-plus labeled images of 29 species, with individual species represented by 59 -315 images.
Comparative Analysis of Text Classification Approaches in Electronic Health Records
May 08, 2020
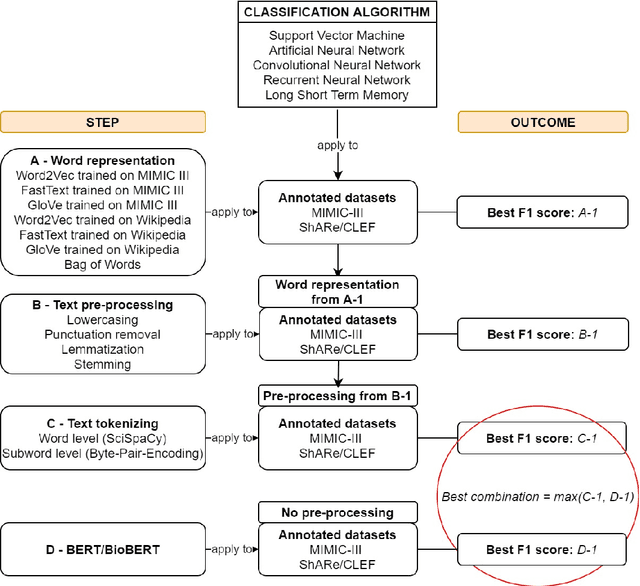

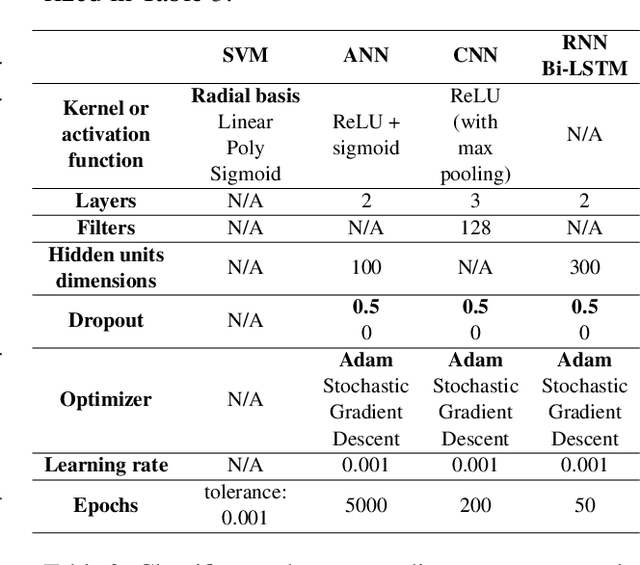
Abstract:Text classification tasks which aim at harvesting and/or organizing information from electronic health records are pivotal to support clinical and translational research. However these present specific challenges compared to other classification tasks, notably due to the particular nature of the medical lexicon and language used in clinical records. Recent advances in embedding methods have shown promising results for several clinical tasks, yet there is no exhaustive comparison of such approaches with other commonly used word representations and classification models. In this work, we analyse the impact of various word representations, text pre-processing and classification algorithms on the performance of four different text classification tasks. The results show that traditional approaches, when tailored to the specific language and structure of the text inherent to the classification task, can achieve or exceed the performance of more recent ones based on contextual embeddings such as BERT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge