Inwan Yoo
PseudoEdgeNet: Nuclei Segmentation only with Point Annotations
Jul 22, 2019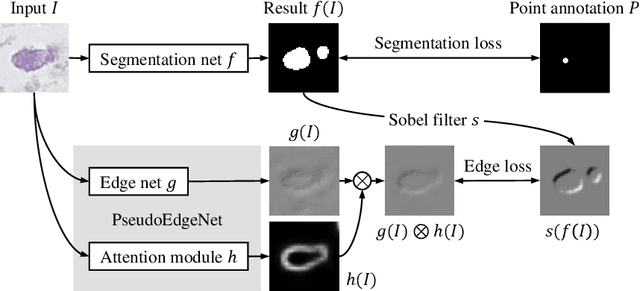
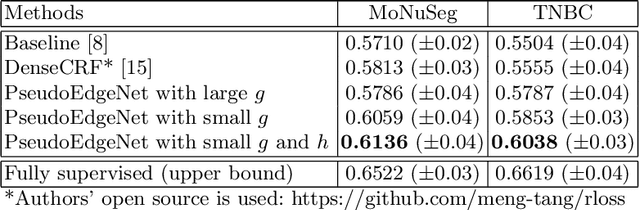
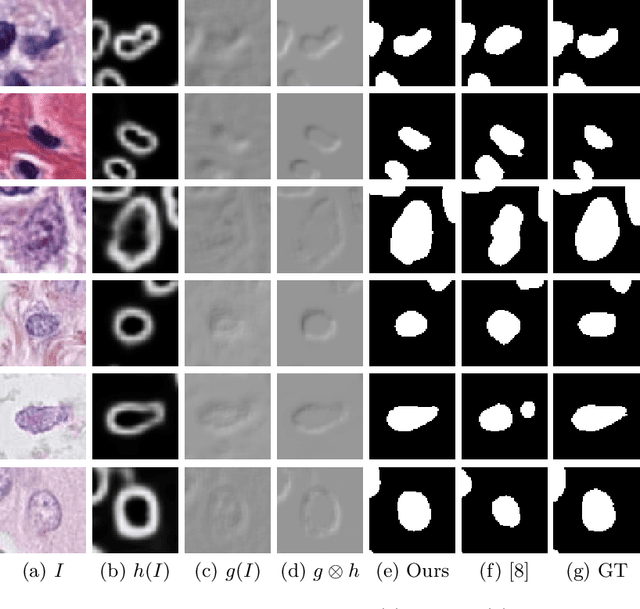
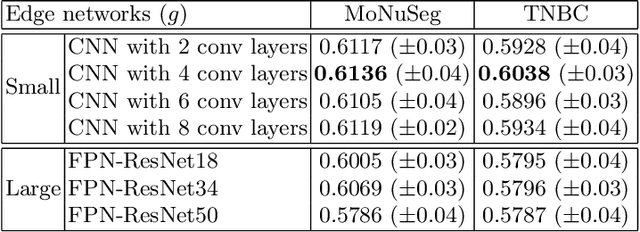
Abstract:Nuclei segmentation is one of the important tasks for whole slide image analysis in digital pathology. With the drastic advance of deep learning, recent deep networks have demonstrated successful performance of the nuclei segmentation task. However, a major bottleneck to achieving good performance is the cost for annotation. A large network requires a large number of segmentation masks, and this annotation task is given to pathologists, not the public. In this paper, we propose a weakly supervised nuclei segmentation method, which requires only point annotations for training. This method can scale to large training set as marking a point of a nucleus is much cheaper than the fine segmentation mask. To this end, we introduce a novel auxiliary network, called PseudoEdgeNet, which guides the segmentation network to recognize nuclei edges even without edge annotations. We evaluate our method with two public datasets, and the results demonstrate that the method consistently outperforms other weakly supervised methods.
ssEMnet: Serial-section Electron Microscopy Image Registration using a Spatial Transformer Network with Learned Features
Dec 05, 2017



Abstract:The alignment of serial-section electron microscopy (ssEM) images is critical for efforts in neuroscience that seek to reconstruct neuronal circuits. However, each ssEM plane contains densely packed structures that vary from one section to the next, which makes matching features across images a challenge. Advances in deep learning has resulted in unprecedented performance in similar computer vision problems, but to our knowledge, they have not been successfully applied to ssEM image co-registration. In this paper, we introduce a novel deep network model that combines a spatial transformer for image deformation and a convolutional autoencoder for unsupervised feature learning for robust ssEM image alignment. This results in improved accuracy and robustness while requiring substantially less user intervention than conventional methods. We evaluate our method by comparing registration quality across several datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge