Wei-Chung Allen Lee
X-Ray2EM: Uncertainty-Aware Cross-Modality Image Reconstruction from X-Ray to Electron Microscopy in Connectomics
Mar 02, 2023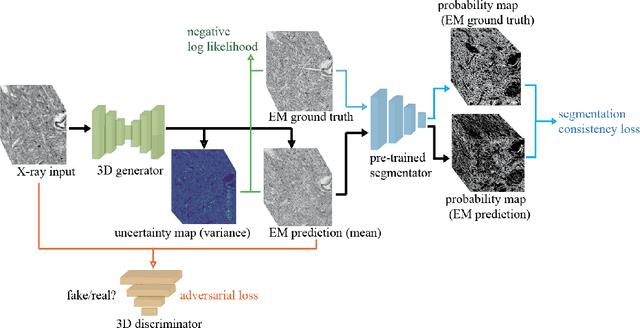
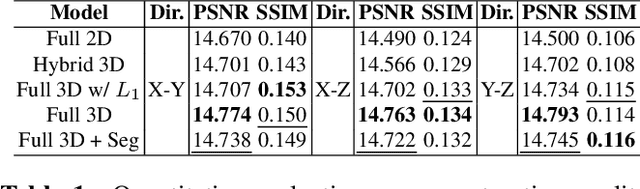
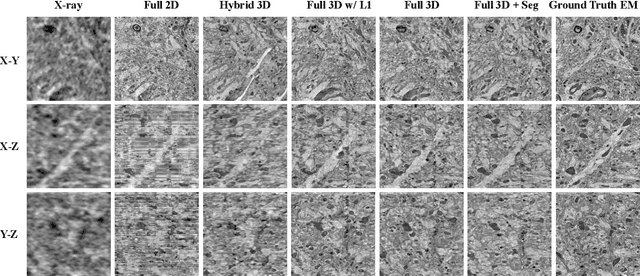
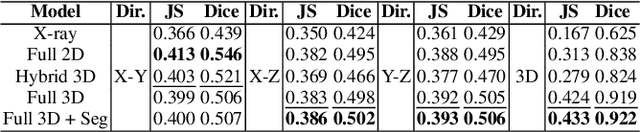
Abstract:Comprehensive, synapse-resolution imaging of the brain will be crucial for understanding neuronal computations and function. In connectomics, this has been the sole purview of volume electron microscopy (EM), which entails an excruciatingly difficult process because it requires cutting tissue into many thin, fragile slices that then need to be imaged, aligned, and reconstructed. Unlike EM, hard X-ray imaging is compatible with thick tissues, eliminating the need for thin sectioning, and delivering fast acquisition, intrinsic alignment, and isotropic resolution. Unfortunately, current state-of-the-art X-ray microscopy provides much lower resolution, to the extent that segmenting membranes is very challenging. We propose an uncertainty-aware 3D reconstruction model that translates X-ray images to EM-like images with enhanced membrane segmentation quality, showing its potential for developing simpler, faster, and more accurate X-ray based connectomics pipelines.
ssEMnet: Serial-section Electron Microscopy Image Registration using a Spatial Transformer Network with Learned Features
Dec 05, 2017



Abstract:The alignment of serial-section electron microscopy (ssEM) images is critical for efforts in neuroscience that seek to reconstruct neuronal circuits. However, each ssEM plane contains densely packed structures that vary from one section to the next, which makes matching features across images a challenge. Advances in deep learning has resulted in unprecedented performance in similar computer vision problems, but to our knowledge, they have not been successfully applied to ssEM image co-registration. In this paper, we introduce a novel deep network model that combines a spatial transformer for image deformation and a convolutional autoencoder for unsupervised feature learning for robust ssEM image alignment. This results in improved accuracy and robustness while requiring substantially less user intervention than conventional methods. We evaluate our method by comparing registration quality across several datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge