David G. C. Hildebrand
ssEMnet: Serial-section Electron Microscopy Image Registration using a Spatial Transformer Network with Learned Features
Dec 05, 2017



Abstract:The alignment of serial-section electron microscopy (ssEM) images is critical for efforts in neuroscience that seek to reconstruct neuronal circuits. However, each ssEM plane contains densely packed structures that vary from one section to the next, which makes matching features across images a challenge. Advances in deep learning has resulted in unprecedented performance in similar computer vision problems, but to our knowledge, they have not been successfully applied to ssEM image co-registration. In this paper, we introduce a novel deep network model that combines a spatial transformer for image deformation and a convolutional autoencoder for unsupervised feature learning for robust ssEM image alignment. This results in improved accuracy and robustness while requiring substantially less user intervention than conventional methods. We evaluate our method by comparing registration quality across several datasets.
Finding Mirror Symmetry via Registration
Mar 31, 2017



Abstract:Symmetry is prevalent in nature and a common theme in man-made designs. Both the human visual system and computer vision algorithms can use symmetry to facilitate object recognition and other tasks. Detecting mirror symmetry in images and data is, therefore, useful for a number of applications. Here, we demonstrate that the problem of fitting a plane of mirror symmetry to data in any Euclidian space can be reduced to the problem of registering two datasets. The exactness of the resulting solution depends entirely on the registration accuracy. This new Mirror Symmetry via Registration (MSR) framework involves (1) data reflection with respect to an arbitrary plane, (2) registration of original and reflected datasets, and (3) calculation of the eigenvector of eigenvalue -1 for the transformation matrix representing the reflection and registration mappings. To support MSR, we also introduce a novel 2D registration method based on random sample consensus of an ensemble of normalized cross-correlation matches. With this as its registration back-end, MSR achieves state-of-the-art performance for symmetry line detection in two independent 2D testing databases. We further demonstrate the generality of MSR by testing it on a database of 3D shapes with an iterative closest point registration back-end. Finally, we explore its applicability to examining symmetry in natural systems by assessing the degree of symmetry present in myelinated axon reconstructions from a larval zebrafish.
FusionNet: A deep fully residual convolutional neural network for image segmentation in connectomics
Dec 26, 2016



Abstract:Electron microscopic connectomics is an ambitious research direction with the goal of studying comprehensive brain connectivity maps by using high-throughput, nano-scale microscopy. One of the main challenges in connectomics research is developing scalable image analysis algorithms that require minimal user intervention. Recently, deep learning has drawn much attention in computer vision because of its exceptional performance in image classification tasks. For this reason, its application to connectomic analyses holds great promise, as well. In this paper, we introduce a novel deep neural network architecture, FusionNet, for the automatic segmentation of neuronal structures in connectomics data. FusionNet leverages the latest advances in machine learning, such as semantic segmentation and residual neural networks, with the novel introduction of summation-based skip connections to allow a much deeper network architecture for a more accurate segmentation. We demonstrate the performance of the proposed method by comparing it with state-of-the-art electron microscopy (EM) segmentation methods from the ISBI EM segmentation challenge. We also show the segmentation results on two different tasks including cell membrane and cell body segmentation and a statistical analysis of cell morphology.
Registering large volume serial-section electron microscopy image sets for neural circuit reconstruction using FFT signal whitening
Dec 14, 2016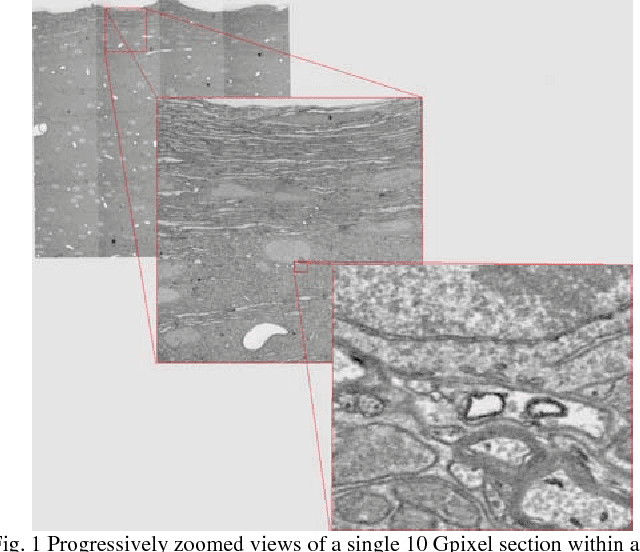
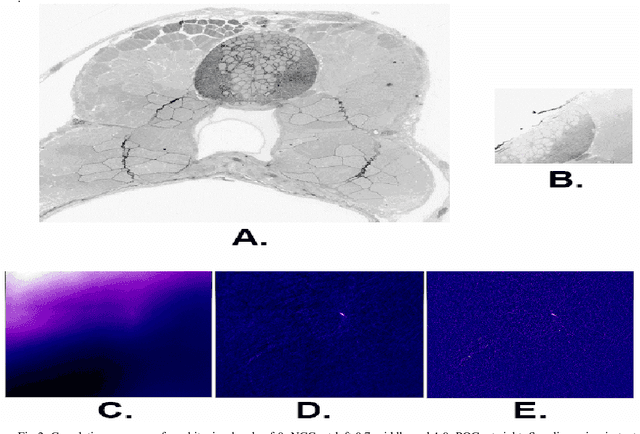
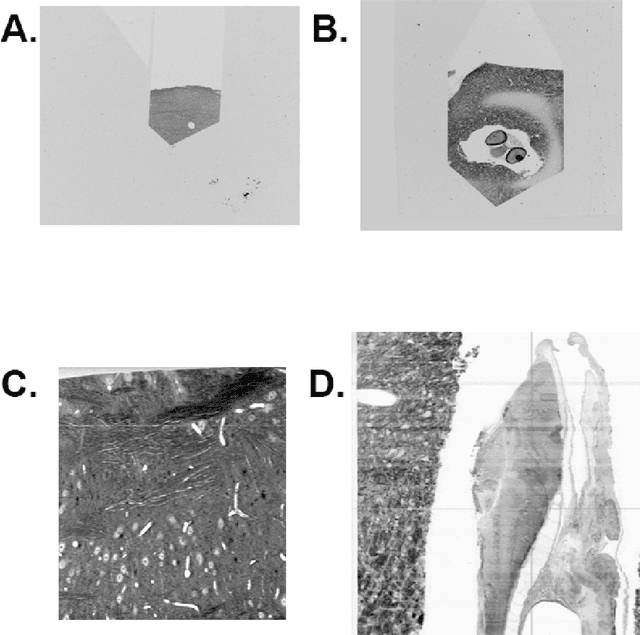
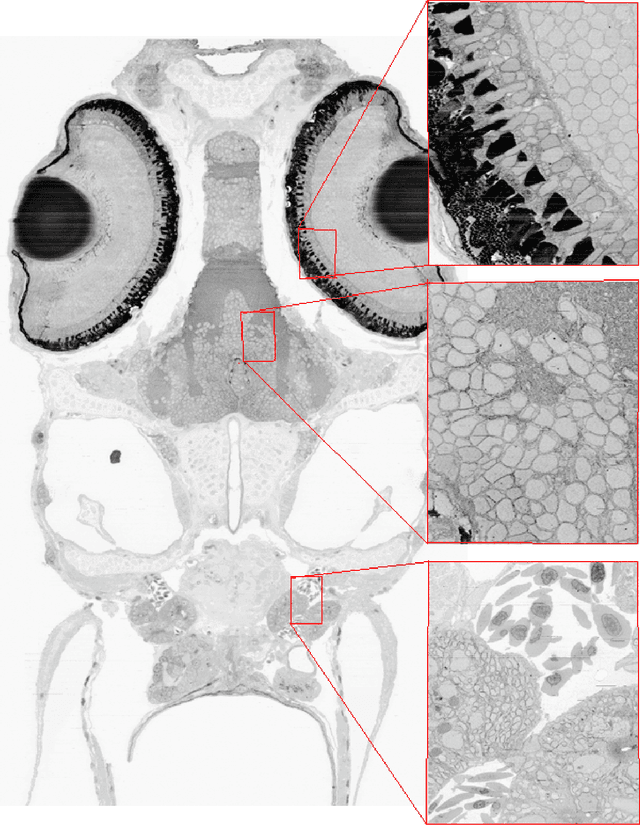
Abstract:The detailed reconstruction of neural anatomy for connectomics studies requires a combination of resolution and large three-dimensional data capture provided by serial section electron microscopy (ssEM). The convergence of high throughput ssEM imaging and improved tissue preparation methods now allows ssEM capture of complete specimen volumes up to cubic millimeter scale. The resulting multi-terabyte image sets span thousands of serial sections and must be precisely registered into coherent volumetric forms in which neural circuits can be traced and segmented. This paper introduces a Signal Whitening Fourier Transform Image Registration approach (SWiFT-IR) under development at the Pittsburgh Supercomputing Center and its use to align mouse and zebrafish brain datasets acquired using the wafer mapper ssEM imaging technology recently developed at Harvard University. Unlike other methods now used for ssEM registration, SWiFT-IR modifies its spatial frequency response during image matching to maximize a signal-to-noise measure used as its primary indicator of alignment quality. This alignment signal is more robust to rapid variations in biological content and unavoidable data distortions than either phase-only or standard Pearson correlation, thus allowing more precise alignment and statistical confidence. These improvements in turn enable an iterative registration procedure based on projections through multiple sections rather than more typical adjacent-pair matching methods. This projection approach, when coupled with known anatomical constraints and iteratively applied in a multi-resolution pyramid fashion, drives the alignment into a smooth form that properly represents complex and widely varying anatomical content such as the full cross-section zebrafish data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge