Registering large volume serial-section electron microscopy image sets for neural circuit reconstruction using FFT signal whitening
Paper and Code
Dec 14, 2016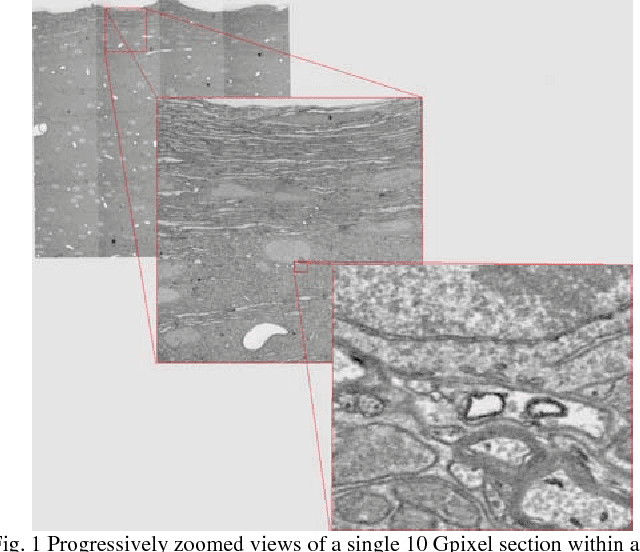
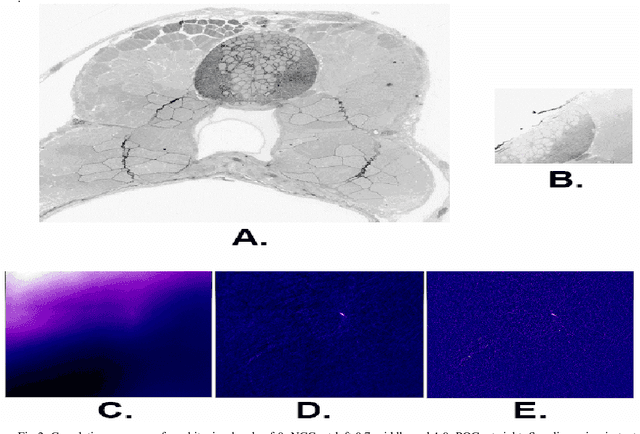
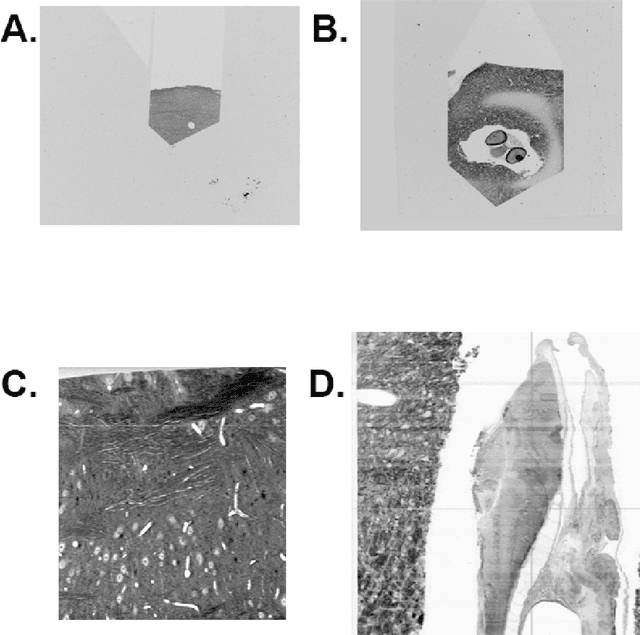
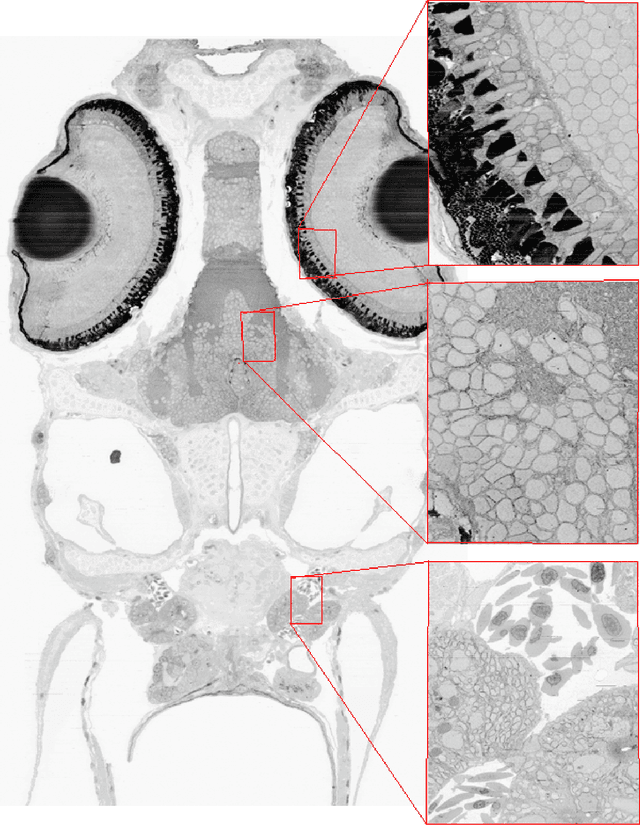
The detailed reconstruction of neural anatomy for connectomics studies requires a combination of resolution and large three-dimensional data capture provided by serial section electron microscopy (ssEM). The convergence of high throughput ssEM imaging and improved tissue preparation methods now allows ssEM capture of complete specimen volumes up to cubic millimeter scale. The resulting multi-terabyte image sets span thousands of serial sections and must be precisely registered into coherent volumetric forms in which neural circuits can be traced and segmented. This paper introduces a Signal Whitening Fourier Transform Image Registration approach (SWiFT-IR) under development at the Pittsburgh Supercomputing Center and its use to align mouse and zebrafish brain datasets acquired using the wafer mapper ssEM imaging technology recently developed at Harvard University. Unlike other methods now used for ssEM registration, SWiFT-IR modifies its spatial frequency response during image matching to maximize a signal-to-noise measure used as its primary indicator of alignment quality. This alignment signal is more robust to rapid variations in biological content and unavoidable data distortions than either phase-only or standard Pearson correlation, thus allowing more precise alignment and statistical confidence. These improvements in turn enable an iterative registration procedure based on projections through multiple sections rather than more typical adjacent-pair matching methods. This projection approach, when coupled with known anatomical constraints and iteratively applied in a multi-resolution pyramid fashion, drives the alignment into a smooth form that properly represents complex and widely varying anatomical content such as the full cross-section zebrafish data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge