Tran Minh Quan
Denoising Diffusion Medical Models
Apr 19, 2023Abstract:In this study, we introduce a generative model that can synthesize a large number of radiographical image/label pairs, and thus is asymptotically favorable to downstream activities such as segmentation in bio-medical image analysis. Denoising Diffusion Medical Model (DDMM), the proposed technique, can create realistic X-ray images and associated segmentations on a small number of annotated datasets as well as other massive unlabeled datasets with no supervision. Radiograph/segmentation pairs are generated jointly by the DDMM sampling process in probabilistic mode. As a result, a vanilla UNet that uses this data augmentation for segmentation task outperforms other similarly data-centric approaches.
Neural Radiance Projection
Mar 15, 2022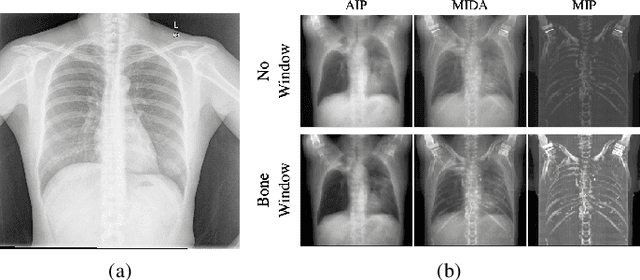
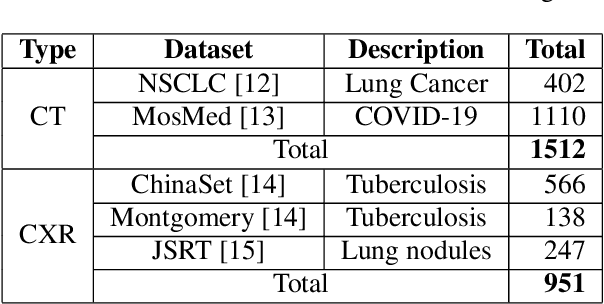
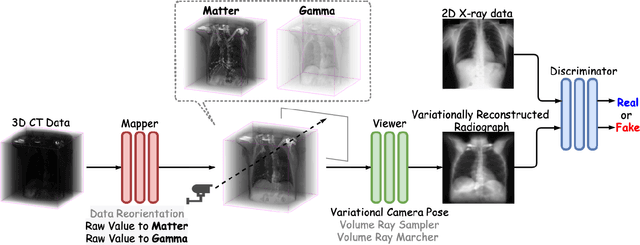
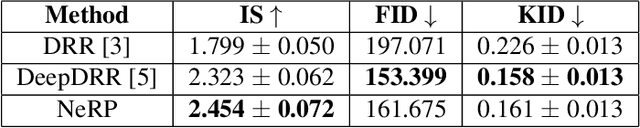
Abstract:The proposed method, Neural Radiance Projection (NeRP), addresses the three most fundamental shortages of training such a convolutional neural network on X-ray image segmentation: dealing with missing/limited human-annotated datasets; ambiguity on the per-pixel label; and the imbalance across positive- and negative- classes distribution. By harnessing a generative adversarial network, we can synthesize a massive amount of physics-based X-ray images, so-called Variationally Reconstructed Radiographs (VRRs), alongside their segmentation from more accurate labeled 3D Computed Tomography data. As a result, VRRs present more faithfully than other projection methods in terms of photo-realistic metrics. Adding outputs from NeRP also surpasses the vanilla UNet models trained on the same pairs of X-ray images.
Reinforced Coloring for End-to-End Instance Segmentation
May 19, 2020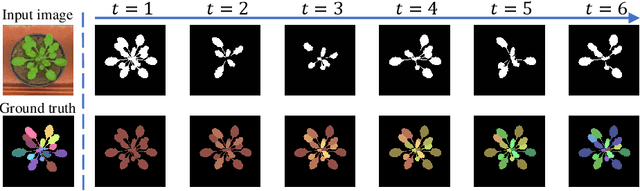
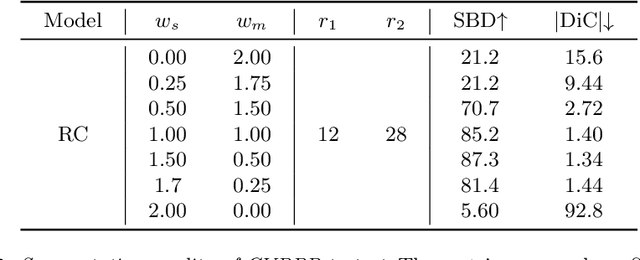
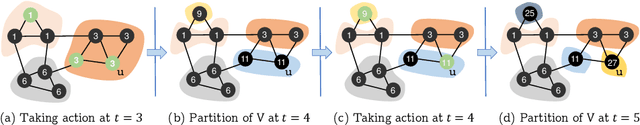
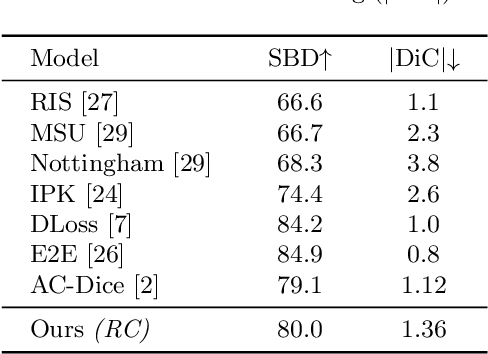
Abstract:Instance segmentation is one of the actively studied research topics in computer vision in which many objects of interest should be separated individually. While many feed-forward networks produce high-quality segmentation on different types of images, their results often suffer from topological errors (merging or splitting) for segmentation of many objects, requiring post-processing. Existing iterative methods, on the other hand, extract a single object at a time using discriminative knowledge-based properties (shapes, boundaries, etc.) without relying on post-processing, but they do not scale well. To exploit the advantages of conventional single-object-per-step segmentation methods without impairing the scalability, we propose a novel iterative deep reinforcement learning agent that learns how to differentiate multiple objects in parallel. Our reward function for the trainable agent is designed to favor grouping pixels belonging to the same object using a graph coloring algorithm. We demonstrate that the proposed method can efficiently perform instance segmentation of many objects without heavy post-processing.
Compressed Sensing MRI Reconstruction using a Generative Adversarial Network with a Cyclic Loss
Mar 15, 2018



Abstract:Compressed Sensing MRI (CS-MRI) has provided theoretical foundations upon which the time-consuming MRI acquisition process can be accelerated. However, it primarily relies on iterative numerical solvers which still hinders their adaptation in time-critical applications. In addition, recent advances in deep neural networks have shown their potential in computer vision and image processing, but their adaptation to MRI reconstruction is still in an early stage. In this paper, we propose a novel deep learning-based generative adversarial model, RefineGAN, for fast and accurate CS-MRI reconstruction. The proposed model is a variant of fully-residual convolutional autoencoder and generative adversarial networks (GANs), specifically designed for CS-MRI formulation; it employs deeper generator and discriminator networks with cyclic data consistency loss for faithful interpolation in the given under-sampled k-space data. In addition, our solution leverages a chained network to further enhance the reconstruction quality. RefineGAN is fast and accurate -- the reconstruction process is extremely rapid, as low as tens of milliseconds for reconstruction of a 256x256 image, because it is one-way deployment on a feed-forward network, and the image quality is superior even for extremely low sampling rate (as low as 10%) due to the data-driven nature of the method. We demonstrate that RefineGAN outperforms the state-of-the-art CS-MRI methods by a large margin in terms of both running time and image quality via evaluation using several open-source MRI databases.
* submitted to IEEE Transactions on Medical Imaging
FusionNet: A deep fully residual convolutional neural network for image segmentation in connectomics
Dec 26, 2016



Abstract:Electron microscopic connectomics is an ambitious research direction with the goal of studying comprehensive brain connectivity maps by using high-throughput, nano-scale microscopy. One of the main challenges in connectomics research is developing scalable image analysis algorithms that require minimal user intervention. Recently, deep learning has drawn much attention in computer vision because of its exceptional performance in image classification tasks. For this reason, its application to connectomic analyses holds great promise, as well. In this paper, we introduce a novel deep neural network architecture, FusionNet, for the automatic segmentation of neuronal structures in connectomics data. FusionNet leverages the latest advances in machine learning, such as semantic segmentation and residual neural networks, with the novel introduction of summation-based skip connections to allow a much deeper network architecture for a more accurate segmentation. We demonstrate the performance of the proposed method by comparing it with state-of-the-art electron microscopy (EM) segmentation methods from the ISBI EM segmentation challenge. We also show the segmentation results on two different tasks including cell membrane and cell body segmentation and a statistical analysis of cell morphology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge