Lu Mi
Mind Meets Space: Rethinking Agentic Spatial Intelligence from a Neuroscience-inspired Perspective
Sep 11, 2025Abstract:Recent advances in agentic AI have led to systems capable of autonomous task execution and language-based reasoning, yet their spatial reasoning abilities remain limited and underexplored, largely constrained to symbolic and sequential processing. In contrast, human spatial intelligence, rooted in integrated multisensory perception, spatial memory, and cognitive maps, enables flexible, context-aware decision-making in unstructured environments. Therefore, bridging this gap is critical for advancing Agentic Spatial Intelligence toward better interaction with the physical 3D world. To this end, we first start from scrutinizing the spatial neural models as studied in computational neuroscience, and accordingly introduce a novel computational framework grounded in neuroscience principles. This framework maps core biological functions to six essential computation modules: bio-inspired multimodal sensing, multi-sensory integration, egocentric-allocentric conversion, an artificial cognitive map, spatial memory, and spatial reasoning. Together, these modules form a perspective landscape for agentic spatial reasoning capability across both virtual and physical environments. On top, we conduct a framework-guided analysis of recent methods, evaluating their relevance to each module and identifying critical gaps that hinder the development of more neuroscience-grounded spatial reasoning modules. We further examine emerging benchmarks and datasets and explore potential application domains ranging from virtual to embodied systems, such as robotics. Finally, we outline potential research directions, emphasizing the promising roadmap that can generalize spatial reasoning across dynamic or unstructured environments. We hope this work will benefit the research community with a neuroscience-grounded perspective and a structured pathway. Our project page can be found at Github.
Concept-Based Unsupervised Domain Adaptation
May 08, 2025Abstract:Concept Bottleneck Models (CBMs) enhance interpretability by explaining predictions through human-understandable concepts but typically assume that training and test data share the same distribution. This assumption often fails under domain shifts, leading to degraded performance and poor generalization. To address these limitations and improve the robustness of CBMs, we propose the Concept-based Unsupervised Domain Adaptation (CUDA) framework. CUDA is designed to: (1) align concept representations across domains using adversarial training, (2) introduce a relaxation threshold to allow minor domain-specific differences in concept distributions, thereby preventing performance drop due to over-constraints of these distributions, (3) infer concepts directly in the target domain without requiring labeled concept data, enabling CBMs to adapt to diverse domains, and (4) integrate concept learning into conventional domain adaptation (DA) with theoretical guarantees, improving interpretability and establishing new benchmarks for DA. Experiments demonstrate that our approach significantly outperforms the state-of-the-art CBM and DA methods on real-world datasets.
Active learning of neural population dynamics using two-photon holographic optogenetics
Dec 03, 2024



Abstract:Recent advances in techniques for monitoring and perturbing neural populations have greatly enhanced our ability to study circuits in the brain. In particular, two-photon holographic optogenetics now enables precise photostimulation of experimenter-specified groups of individual neurons, while simultaneous two-photon calcium imaging enables the measurement of ongoing and induced activity across the neural population. Despite the enormous space of potential photostimulation patterns and the time-consuming nature of photostimulation experiments, very little algorithmic work has been done to determine the most effective photostimulation patterns for identifying the neural population dynamics. Here, we develop methods to efficiently select which neurons to stimulate such that the resulting neural responses will best inform a dynamical model of the neural population activity. Using neural population responses to photostimulation in mouse motor cortex, we demonstrate the efficacy of a low-rank linear dynamical systems model, and develop an active learning procedure which takes advantage of low-rank structure to determine informative photostimulation patterns. We demonstrate our approach on both real and synthetic data, obtaining in some cases as much as a two-fold reduction in the amount of data required to reach a given predictive power. Our active stimulation design method is based on a novel active learning procedure for low-rank regression, which may be of independent interest.
Energy-Based Concept Bottleneck Models: Unifying Prediction, Concept Intervention, and Conditional Interpretations
Jan 25, 2024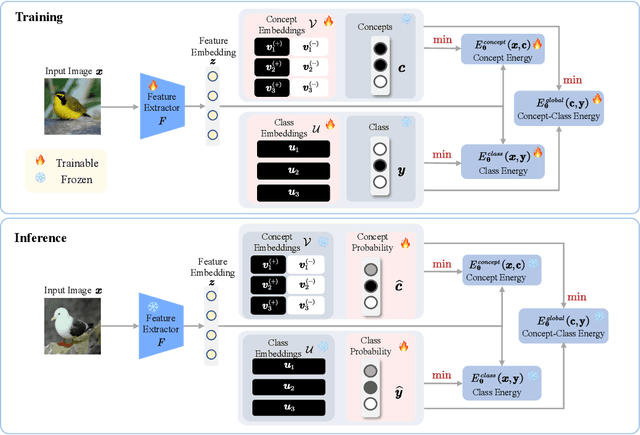

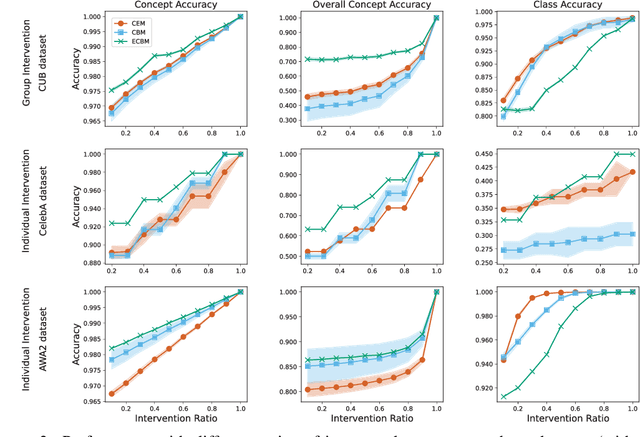

Abstract:Existing methods, such as concept bottleneck models (CBMs), have been successful in providing concept-based interpretations for black-box deep learning models. They typically work by predicting concepts given the input and then predicting the final class label given the predicted concepts. However, (1) they often fail to capture the high-order, nonlinear interaction between concepts, e.g., correcting a predicted concept (e.g., "yellow breast") does not help correct highly correlated concepts (e.g., "yellow belly"), leading to suboptimal final accuracy; (2) they cannot naturally quantify the complex conditional dependencies between different concepts and class labels (e.g., for an image with the class label "Kentucky Warbler" and a concept "black bill", what is the probability that the model correctly predicts another concept "black crown"), therefore failing to provide deeper insight into how a black-box model works. In response to these limitations, we propose Energy-based Concept Bottleneck Models (ECBMs). Our ECBMs use a set of neural networks to define the joint energy of candidate (input, concept, class) tuples. With such a unified interface, prediction, concept correction, and conditional dependency quantification are then represented as conditional probabilities, which are generated by composing different energy functions. Our ECBMs address both limitations of existing CBMs, providing higher accuracy and richer concept interpretations. Empirical results show that our approach outperforms the state-of-the-art on real-world datasets.
Attention for Causal Relationship Discovery from Biological Neural Dynamics
Nov 23, 2023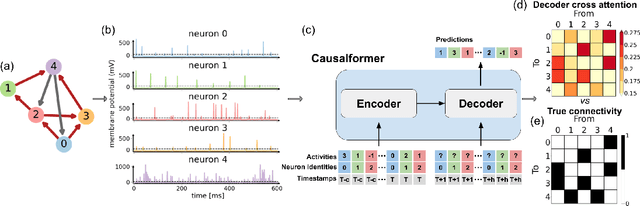
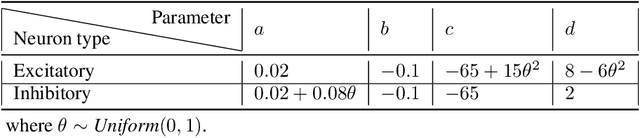
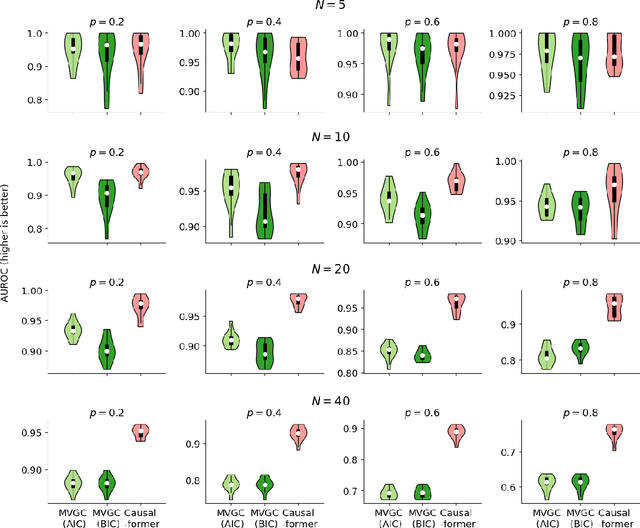
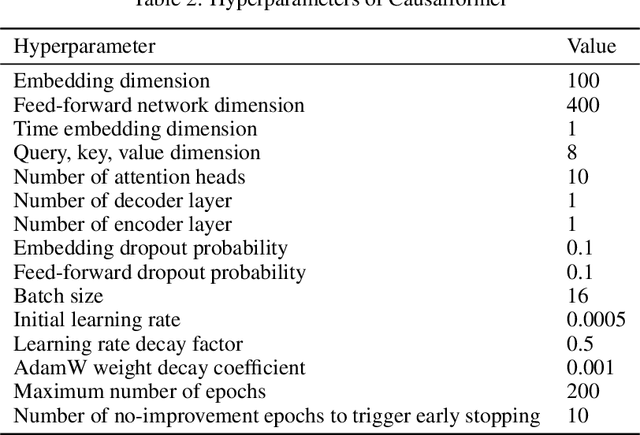
Abstract:This paper explores the potential of the transformer models for learning Granger causality in networks with complex nonlinear dynamics at every node, as in neurobiological and biophysical networks. Our study primarily focuses on a proof-of-concept investigation based on simulated neural dynamics, for which the ground-truth causality is known through the underlying connectivity matrix. For transformer models trained to forecast neuronal population dynamics, we show that the cross attention module effectively captures the causal relationship among neurons, with an accuracy equal or superior to that for the most popular Granger causality analysis method. While we acknowledge that real-world neurobiology data will bring further challenges, including dynamic connectivity and unobserved variability, this research offers an encouraging preliminary glimpse into the utility of the transformer model for causal representation learning in neuroscience.
Learning Time-Invariant Representations for Individual Neurons from Population Dynamics
Nov 03, 2023



Abstract:Neurons can display highly variable dynamics. While such variability presumably supports the wide range of behaviors generated by the organism, their gene expressions are relatively stable in the adult brain. This suggests that neuronal activity is a combination of its time-invariant identity and the inputs the neuron receives from the rest of the circuit. Here, we propose a self-supervised learning based method to assign time-invariant representations to individual neurons based on permutation-, and population size-invariant summary of population recordings. We fit dynamical models to neuronal activity to learn a representation by considering the activity of both the individual and the neighboring population. Our self-supervised approach and use of implicit representations enable robust inference against imperfections such as partial overlap of neurons across sessions, trial-to-trial variability, and limited availability of molecular (transcriptomic) labels for downstream supervised tasks. We demonstrate our method on a public multimodal dataset of mouse cortical neuronal activity and transcriptomic labels. We report > 35% improvement in predicting the transcriptomic subclass identity and > 20% improvement in predicting class identity with respect to the state-of-the-art.
LatticeGen: A Cooperative Framework which Hides Generated Text in a Lattice for Privacy-Aware Generation on Cloud
Oct 02, 2023



Abstract:In the current user-server interaction paradigm of prompted generation with large language models (LLM) on cloud, the server fully controls the generation process, which leaves zero options for users who want to keep the generated text to themselves. We propose LatticeGen, a cooperative framework in which the server still handles most of the computation while the user controls the sampling operation. The key idea is that the true generated sequence is mixed with noise tokens by the user and hidden in a noised lattice. Considering potential attacks from a hypothetically malicious server and how the user can defend against it, we propose the repeated beam-search attack and the mixing noise scheme. In our experiments we apply LatticeGen to protect both prompt and generation. It is shown that while the noised lattice degrades generation quality, LatticeGen successfully protects the true generation to a remarkable degree under strong attacks (more than 50% of the semantic remains hidden as measured by BERTScore).
X-Ray2EM: Uncertainty-Aware Cross-Modality Image Reconstruction from X-Ray to Electron Microscopy in Connectomics
Mar 02, 2023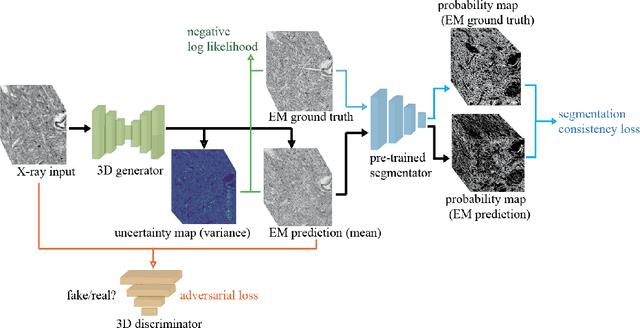
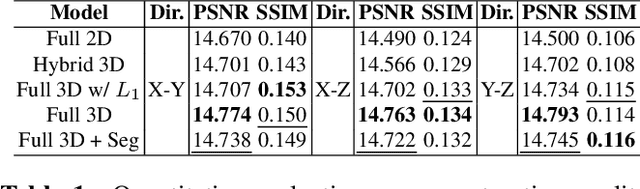
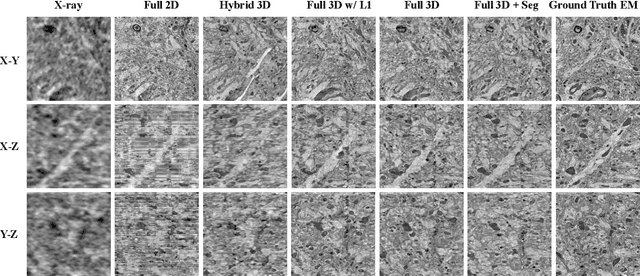
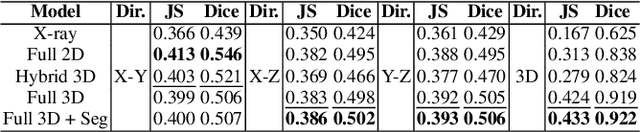
Abstract:Comprehensive, synapse-resolution imaging of the brain will be crucial for understanding neuronal computations and function. In connectomics, this has been the sole purview of volume electron microscopy (EM), which entails an excruciatingly difficult process because it requires cutting tissue into many thin, fragile slices that then need to be imaged, aligned, and reconstructed. Unlike EM, hard X-ray imaging is compatible with thick tissues, eliminating the need for thin sectioning, and delivering fast acquisition, intrinsic alignment, and isotropic resolution. Unfortunately, current state-of-the-art X-ray microscopy provides much lower resolution, to the extent that segmenting membranes is very challenging. We propose an uncertainty-aware 3D reconstruction model that translates X-ray images to EM-like images with enhanced membrane segmentation quality, showing its potential for developing simpler, faster, and more accurate X-ray based connectomics pipelines.
The XPRESS Challenge: Xray Projectomic Reconstruction -- Extracting Segmentation with Skeletons
Feb 24, 2023



Abstract:The wiring and connectivity of neurons form a structural basis for the function of the nervous system. Advances in volume electron microscopy (EM) and image segmentation have enabled mapping of circuit diagrams (connectomics) within local regions of the mouse brain. However, applying volume EM over the whole brain is not currently feasible due to technological challenges. As a result, comprehensive maps of long-range connections between brain regions are lacking. Recently, we demonstrated that X-ray holographic nanotomography (XNH) can provide high-resolution images of brain tissue at a much larger scale than EM. In particular, XNH is wellsuited to resolve large, myelinated axon tracts (white matter) that make up the bulk of long-range connections (projections) and are critical for inter-region communication. Thus, XNH provides an imaging solution for brain-wide projectomics. However, because XNH data is typically collected at lower resolutions and larger fields-of-view than EM, accurate segmentation of XNH images remains an important challenge that we present here. In this task, we provide volumetric XNH images of cortical white matter axons from the mouse brain along with ground truth annotations for axon trajectories. Manual voxel-wise annotation of ground truth is a time-consuming bottleneck for training segmentation networks. On the other hand, skeleton-based ground truth is much faster to annotate, and sufficient to determine connectivity. Therefore, we encourage participants to develop methods to leverage skeleton-based training. To this end, we provide two types of ground-truth annotations: a small volume of voxel-wise annotations and a larger volume with skeleton-based annotations. Entries will be evaluated on how accurately the submitted segmentations agree with the ground-truth skeleton annotations.
im2nerf: Image to Neural Radiance Field in the Wild
Sep 08, 2022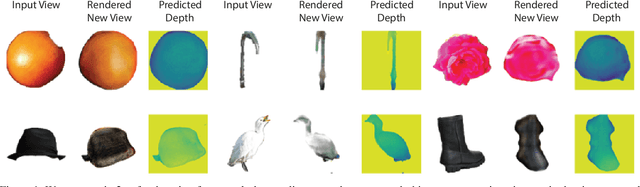
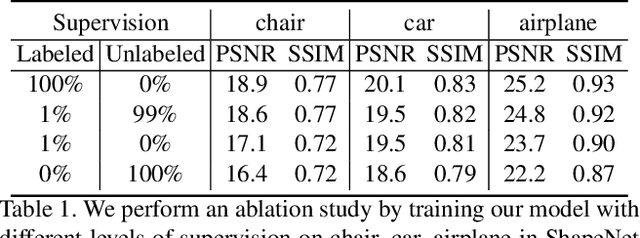
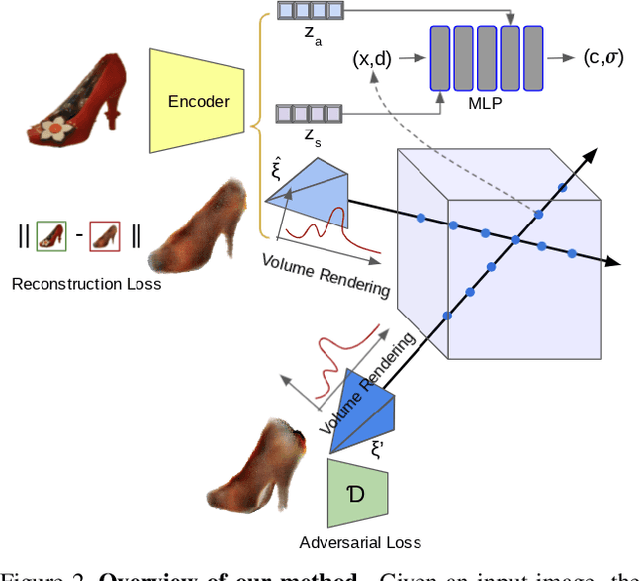
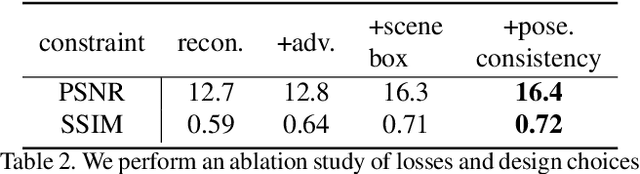
Abstract:We propose im2nerf, a learning framework that predicts a continuous neural object representation given a single input image in the wild, supervised by only segmentation output from off-the-shelf recognition methods. The standard approach to constructing neural radiance fields takes advantage of multi-view consistency and requires many calibrated views of a scene, a requirement that cannot be satisfied when learning on large-scale image data in the wild. We take a step towards addressing this shortcoming by introducing a model that encodes the input image into a disentangled object representation that contains a code for object shape, a code for object appearance, and an estimated camera pose from which the object image is captured. Our model conditions a NeRF on the predicted object representation and uses volume rendering to generate images from novel views. We train the model end-to-end on a large collection of input images. As the model is only provided with single-view images, the problem is highly under-constrained. Therefore, in addition to using a reconstruction loss on the synthesized input view, we use an auxiliary adversarial loss on the novel rendered views. Furthermore, we leverage object symmetry and cycle camera pose consistency. We conduct extensive quantitative and qualitative experiments on the ShapeNet dataset as well as qualitative experiments on Open Images dataset. We show that in all cases, im2nerf achieves the state-of-the-art performance for novel view synthesis from a single-view unposed image in the wild.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge