Zhongren Chen
Quantile-Optimal Policy Learning under Unmeasured Confounding
Jun 08, 2025Abstract:We study quantile-optimal policy learning where the goal is to find a policy whose reward distribution has the largest $\alpha$-quantile for some $\alpha \in (0, 1)$. We focus on the offline setting whose generating process involves unobserved confounders. Such a problem suffers from three main challenges: (i) nonlinearity of the quantile objective as a functional of the reward distribution, (ii) unobserved confounding issue, and (iii) insufficient coverage of the offline dataset. To address these challenges, we propose a suite of causal-assisted policy learning methods that provably enjoy strong theoretical guarantees under mild conditions. In particular, to address (i) and (ii), using causal inference tools such as instrumental variables and negative controls, we propose to estimate the quantile objectives by solving nonlinear functional integral equations. Then we adopt a minimax estimation approach with nonparametric models to solve these integral equations, and propose to construct conservative policy estimates that address (iii). The final policy is the one that maximizes these pessimistic estimates. In addition, we propose a novel regularized policy learning method that is more amenable to computation. Finally, we prove that the policies learned by these methods are $\tilde{\mathscr{O}}(n^{-1/2})$ quantile-optimal under a mild coverage assumption on the offline dataset. Here, $\tilde{\mathscr{O}}(\cdot)$ omits poly-logarithmic factors. To the best of our knowledge, we propose the first sample-efficient policy learning algorithms for estimating the quantile-optimal policy when there exist unmeasured confounding.
A Framework to Assess the Persuasion Risks Large Language Model Chatbots Pose to Democratic Societies
Apr 29, 2025Abstract:In recent years, significant concern has emerged regarding the potential threat that Large Language Models (LLMs) pose to democratic societies through their persuasive capabilities. We expand upon existing research by conducting two survey experiments and a real-world simulation exercise to determine whether it is more cost effective to persuade a large number of voters using LLM chatbots compared to standard political campaign practice, taking into account both the "receive" and "accept" steps in the persuasion process (Zaller 1992). These experiments improve upon previous work by assessing extended interactions between humans and LLMs (instead of using single-shot interactions) and by assessing both short- and long-run persuasive effects (rather than simply asking users to rate the persuasiveness of LLM-produced content). In two survey experiments (N = 10,417) across three distinct political domains, we find that while LLMs are about as persuasive as actual campaign ads once voters are exposed to them, political persuasion in the real-world depends on both exposure to a persuasive message and its impact conditional on exposure. Through simulations based on real-world parameters, we estimate that LLM-based persuasion costs between \$48-\$74 per persuaded voter compared to \$100 for traditional campaign methods, when accounting for the costs of exposure. However, it is currently much easier to scale traditional campaign persuasion methods than LLM-based persuasion. While LLMs do not currently appear to have substantially greater potential for large-scale political persuasion than existing non-LLM methods, this may change as LLM capabilities continue to improve and it becomes easier to scalably encourage exposure to persuasive LLMs.
CausalEGM: a general causal inference framework by encoding generative modeling
Dec 13, 2022



Abstract:Although understanding and characterizing causal effects have become essential in observational studies, it is challenging when the confounders are high-dimensional. In this article, we develop a general framework $\textit{CausalEGM}$ for estimating causal effects by encoding generative modeling, which can be applied in both binary and continuous treatment settings. Under the potential outcome framework with unconfoundedness, we establish a bidirectional transformation between the high-dimensional confounders space and a low-dimensional latent space where the density is known (e.g., multivariate normal distribution). Through this, CausalEGM simultaneously decouples the dependencies of confounders on both treatment and outcome and maps the confounders to the low-dimensional latent space. By conditioning on the low-dimensional latent features, CausalEGM can estimate the causal effect for each individual or the average causal effect within a population. Our theoretical analysis shows that the excess risk for CausalEGM can be bounded through empirical process theory. Under an assumption on encoder-decoder networks, the consistency of the estimate can be guaranteed. In a series of experiments, CausalEGM demonstrates superior performance over existing methods for both binary and continuous treatments. Specifically, we find CausalEGM to be substantially more powerful than competing methods in the presence of large sample sizes and high dimensional confounders. The software of CausalEGM is freely available at https://github.com/SUwonglab/CausalEGM.
Subgraph Frequency Distribution Estimation using Graph Neural Networks
Jul 14, 2022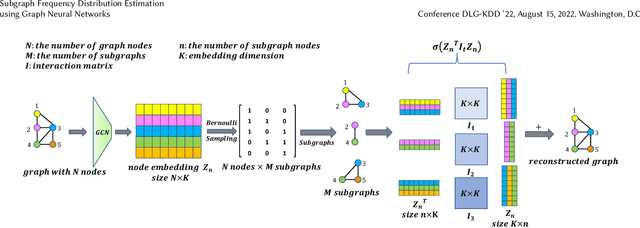
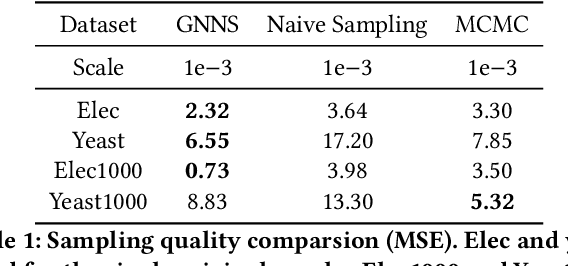
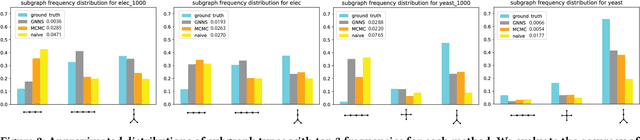
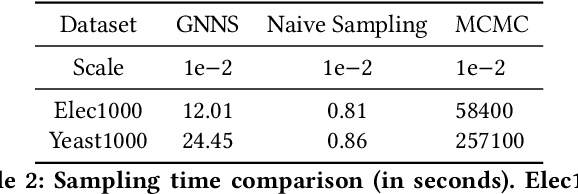
Abstract:Small subgraphs (graphlets) are important features to describe fundamental units of a large network. The calculation of the subgraph frequency distributions has a wide application in multiple domains including biology and engineering. Unfortunately due to the inherent complexity of this task, most of the existing methods are computationally intensive and inefficient. In this work, we propose GNNS, a novel representational learning framework that utilizes graph neural networks to sample subgraphs efficiently for estimating their frequency distribution. Our framework includes an inference model and a generative model that learns hierarchical embeddings of nodes, subgraphs, and graph types. With the learned model and embeddings, subgraphs are sampled in a highly scalable and parallel way and the frequency distribution estimation is then performed based on these sampled subgraphs. Eventually, our methods achieve comparable accuracy and a significant speedup by three orders of magnitude compared to existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge