Anika Tabassum
Training a Custom CNN on Five Heterogeneous Image Datasets
Jan 08, 2026Abstract:Deep learning has transformed visual data analysis, with Convolutional Neural Networks (CNNs) becoming highly effective in learning meaningful feature representations directly from images. Unlike traditional manual feature engineering methods, CNNs automatically extract hierarchical visual patterns, enabling strong performance across diverse real-world contexts. This study investigates the effectiveness of CNN-based architectures across five heterogeneous datasets spanning agricultural and urban domains: mango variety classification, paddy variety identification, road surface condition assessment, auto-rickshaw detection, and footpath encroachment monitoring. These datasets introduce varying challenges, including differences in illumination, resolution, environmental complexity, and class imbalance, necessitating adaptable and robust learning models. We evaluate a lightweight, task-specific custom CNN alongside established deep architectures, including ResNet-18 and VGG-16, trained both from scratch and using transfer learning. Through systematic preprocessing, augmentation, and controlled experimentation, we analyze how architectural complexity, model depth, and pre-training influence convergence, generalization, and performance across datasets of differing scale and difficulty. The key contributions of this work are: (1) the development of an efficient custom CNN that achieves competitive performance across multiple application domains, and (2) a comprehensive comparative analysis highlighting when transfer learning and deep architectures provide substantial advantages, particularly in data-constrained environments. These findings offer practical insights for deploying deep learning models in resource-limited yet high-impact real-world visual classification tasks.
LLM-ProS: Analyzing Large Language Models' Performance in Competitive Problem Solving
Feb 04, 2025Abstract:The rapid advancement of large language models has opened new avenues for automating complex problem-solving tasks such as algorithmic coding and competitive programming. This paper introduces a novel evaluation technique, LLM-ProS, to assess the performance of state-of-the-art LLMs on International Collegiate Programming Contest (ICPC) problems. Using a curated dataset of 166 World Finals problems from 2011 to 2024, we benchmark the models' reasoning, accuracy, and efficiency. We evaluate the five models-GPT-4o, Mistral Large, Llama-3.1-405B, and the o1 family, consisting of o1-mini and o1-preview, across critical metrics like correctness, resource utilization, and response calibration. Our results reveal significant differences in the models' abilities to generalize, adapt, and solve novel problems. We also investigated the impact of training methodologies, dataset contamination, and chain-of-thought reasoning on model performance. The findings provide new insights into optimizing LLMs for algorithmic tasks, highlighting both strengths and limitations of current models.
Adapting Segment Anything Model (SAM) to Experimental Datasets via Fine-Tuning on GAN-based Simulation: A Case Study in Additive Manufacturing
Dec 16, 2024Abstract:Industrial X-ray computed tomography (XCT) is a powerful tool for non-destructive characterization of materials and manufactured components. XCT commonly accompanied by advanced image analysis and computer vision algorithms to extract relevant information from the images. Traditional computer vision models often struggle due to noise, resolution variability, and complex internal structures, particularly in scientific imaging applications. State-of-the-art foundational models, like the Segment Anything Model (SAM)-designed for general-purpose image segmentation-have revolutionized image segmentation across various domains, yet their application in specialized fields like materials science remains under-explored. In this work, we explore the application and limitations of SAM for industrial X-ray CT inspection of additive manufacturing components. We demonstrate that while SAM shows promise, it struggles with out-of-distribution data, multiclass segmentation, and computational efficiency during fine-tuning. To address these issues, we propose a fine-tuning strategy utilizing parameter-efficient techniques, specifically Conv-LoRa, to adapt SAM for material-specific datasets. Additionally, we leverage generative adversarial network (GAN)-generated data to enhance the training process and improve the model's segmentation performance on complex X-ray CT data. Our experimental results highlight the importance of tailored segmentation models for accurate inspection, showing that fine-tuning SAM on domain-specific scientific imaging data significantly improves performance. However, despite improvements, the model's ability to generalize across diverse datasets remains limited, highlighting the need for further research into robust, scalable solutions for domain-specific segmentation tasks.
Reinforcement Learning as a Parsimonious Alternative to Prediction Cascades: A Case Study on Image Segmentation
Feb 19, 2024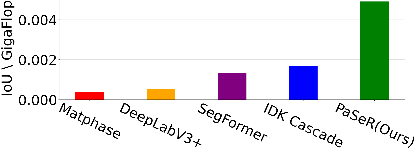

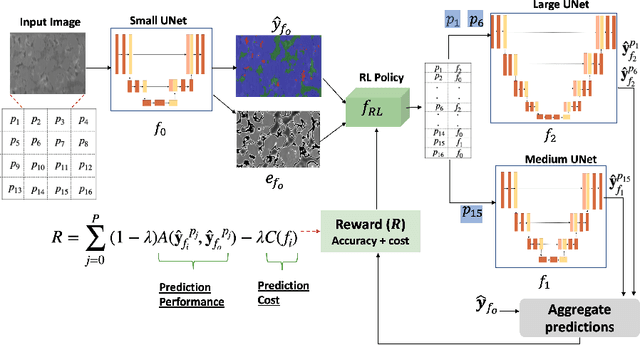

Abstract:Deep learning architectures have achieved state-of-the-art (SOTA) performance on computer vision tasks such as object detection and image segmentation. This may be attributed to the use of over-parameterized, monolithic deep learning architectures executed on large datasets. Although such architectures lead to increased accuracy, this is usually accompanied by a large increase in computation and memory requirements during inference. While this is a non-issue in traditional machine learning pipelines, the recent confluence of machine learning and fields like the Internet of Things has rendered such large architectures infeasible for execution in low-resource settings. In such settings, previous efforts have proposed decision cascades where inputs are passed through models of increasing complexity until desired performance is achieved. However, we argue that cascaded prediction leads to increased computational cost due to wasteful intermediate computations. To address this, we propose PaSeR (Parsimonious Segmentation with Reinforcement Learning) a non-cascading, cost-aware learning pipeline as an alternative to cascaded architectures. Through experimental evaluation on real-world and standard datasets, we demonstrate that PaSeR achieves better accuracy while minimizing computational cost relative to cascaded models. Further, we introduce a new metric IoU/GigaFlop to evaluate the balance between cost and performance. On the real-world task of battery material phase segmentation, PaSeR yields a minimum performance improvement of 174% on the IoU/GigaFlop metric with respect to baselines. We also demonstrate PaSeR's adaptability to complementary models trained on a noisy MNIST dataset, where it achieved a minimum performance improvement on IoU/GigaFlop of 13.4% over SOTA models. Code and data are available at https://github.com/scailab/paser .
Attention for Causal Relationship Discovery from Biological Neural Dynamics
Nov 23, 2023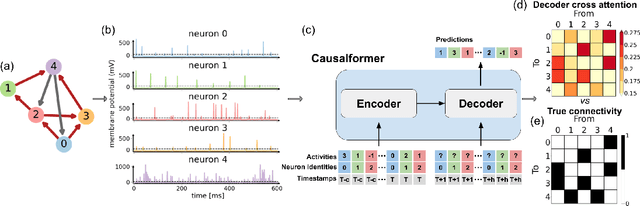
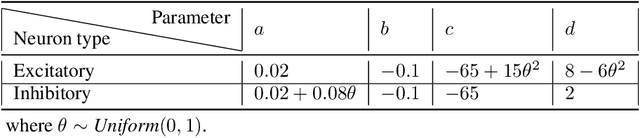
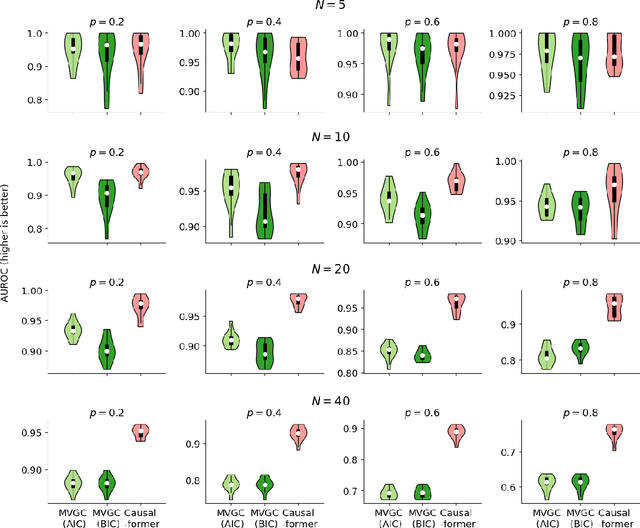
Abstract:This paper explores the potential of the transformer models for learning Granger causality in networks with complex nonlinear dynamics at every node, as in neurobiological and biophysical networks. Our study primarily focuses on a proof-of-concept investigation based on simulated neural dynamics, for which the ground-truth causality is known through the underlying connectivity matrix. For transformer models trained to forecast neuronal population dynamics, we show that the cross attention module effectively captures the causal relationship among neurons, with an accuracy equal or superior to that for the most popular Granger causality analysis method. While we acknowledge that real-world neurobiology data will bring further challenges, including dynamic connectivity and unobserved variability, this research offers an encouraging preliminary glimpse into the utility of the transformer model for causal representation learning in neuroscience.
Snapshot Multispectral Imaging Using a Diffractive Optical Network
Dec 10, 2022Abstract:Multispectral imaging has been used for numerous applications in e.g., environmental monitoring, aerospace, defense, and biomedicine. Here, we present a diffractive optical network-based multispectral imaging system trained using deep learning to create a virtual spectral filter array at the output image field-of-view. This diffractive multispectral imager performs spatially-coherent imaging over a large spectrum, and at the same time, routes a pre-determined set of spectral channels onto an array of pixels at the output plane, converting a monochrome focal plane array or image sensor into a multispectral imaging device without any spectral filters or image recovery algorithms. Furthermore, the spectral responsivity of this diffractive multispectral imager is not sensitive to input polarization states. Through numerical simulations, we present different diffractive network designs that achieve snapshot multispectral imaging with 4, 9 and 16 unique spectral bands within the visible spectrum, based on passive spatially-structured diffractive surfaces, with a compact design that axially spans ~72 times the mean wavelength of the spectral band of interest. Moreover, we experimentally demonstrate a diffractive multispectral imager based on a 3D-printed diffractive network that creates at its output image plane a spatially-repeating virtual spectral filter array with 2x2=4 unique bands at terahertz spectrum. Due to their compact form factor and computation-free, power-efficient and polarization-insensitive forward operation, diffractive multispectral imagers can be transformative for various imaging and sensing applications and be used at different parts of the electromagnetic spectrum where high-density and wide-area multispectral pixel arrays are not widely available.
Diffractive Interconnects: All-Optical Permutation Operation Using Diffractive Networks
Jun 21, 2022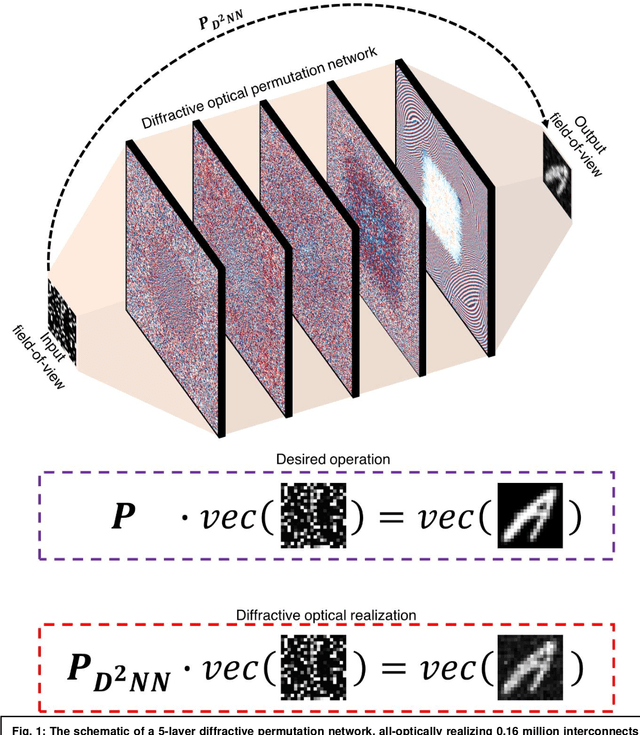
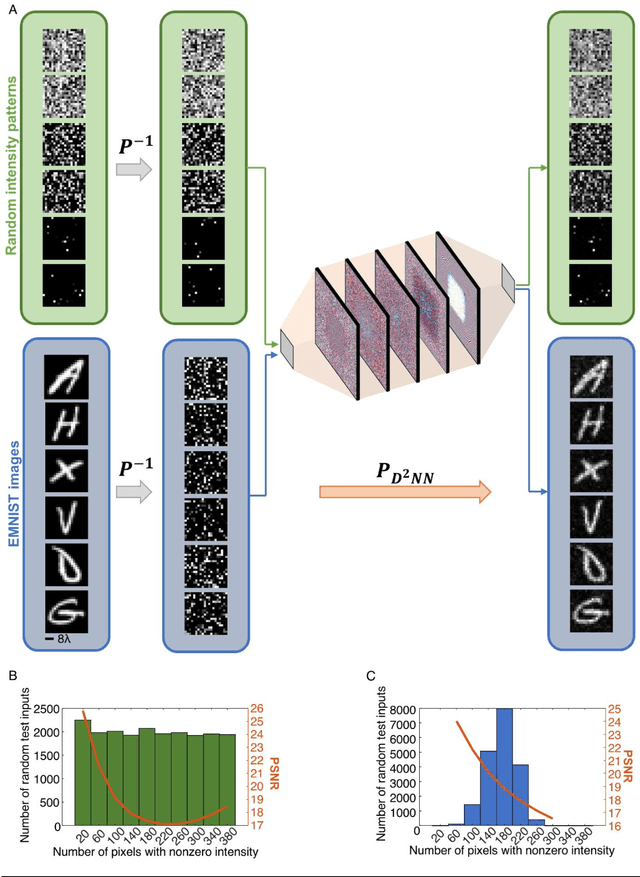
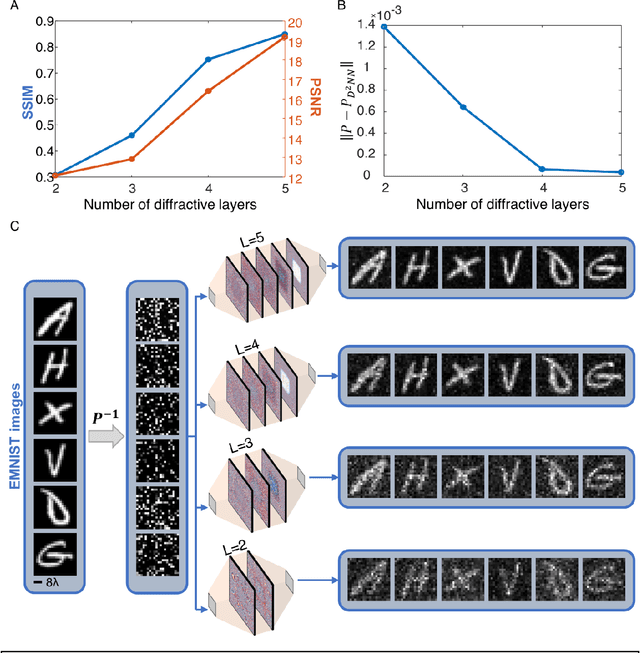
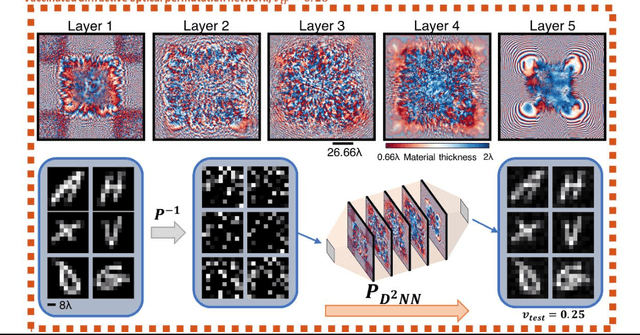
Abstract:Permutation matrices form an important computational building block frequently used in various fields including e.g., communications, information security and data processing. Optical implementation of permutation operators with relatively large number of input-output interconnections based on power-efficient, fast, and compact platforms is highly desirable. Here, we present diffractive optical networks engineered through deep learning to all-optically perform permutation operations that can scale to hundreds of thousands of interconnections between an input and an output field-of-view using passive transmissive layers that are individually structured at the wavelength scale. Our findings indicate that the capacity of the diffractive optical network in approximating a given permutation operation increases proportional to the number of diffractive layers and trainable transmission elements in the system. Such deeper diffractive network designs can pose practical challenges in terms of physical alignment and output diffraction efficiency of the system. We addressed these challenges by designing misalignment tolerant diffractive designs that can all-optically perform arbitrarily-selected permutation operations, and experimentally demonstrated, for the first time, a diffractive permutation network that operates at THz part of the spectrum. Diffractive permutation networks might find various applications in e.g., security, image encryption and data processing, along with telecommunications; especially with the carrier frequencies in wireless communications approaching THz-bands, the presented diffractive permutation networks can potentially serve as channel routing and interconnection panels in wireless networks.
Super-resolution image display using diffractive decoders
Jun 15, 2022
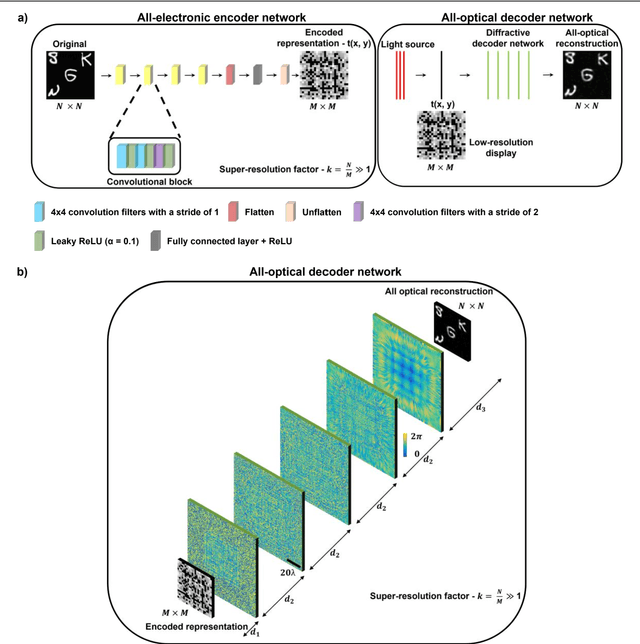
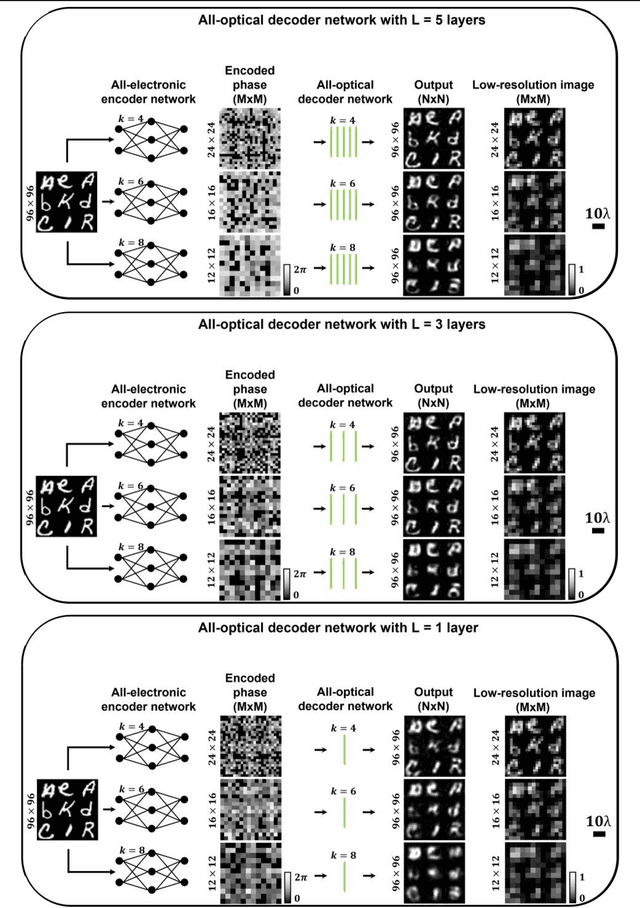
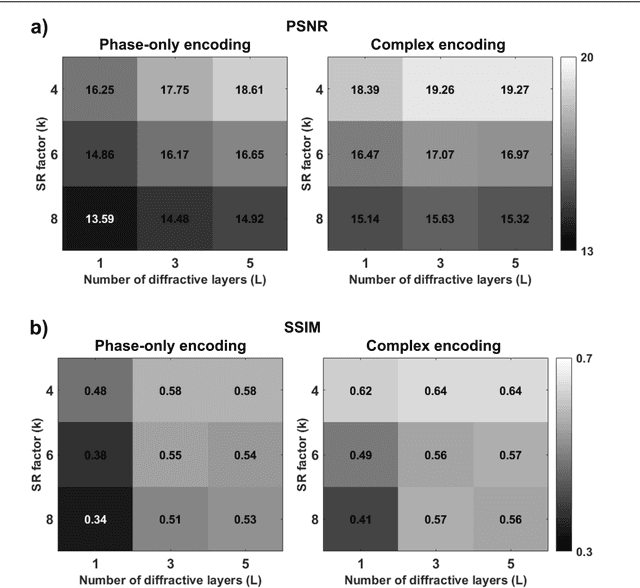
Abstract:High-resolution synthesis/projection of images over a large field-of-view (FOV) is hindered by the restricted space-bandwidth-product (SBP) of wavefront modulators. We report a deep learning-enabled diffractive display design that is based on a jointly-trained pair of an electronic encoder and a diffractive optical decoder to synthesize/project super-resolved images using low-resolution wavefront modulators. The digital encoder, composed of a trained convolutional neural network (CNN), rapidly pre-processes the high-resolution images of interest so that their spatial information is encoded into low-resolution (LR) modulation patterns, projected via a low SBP wavefront modulator. The diffractive decoder processes this LR encoded information using thin transmissive layers that are structured using deep learning to all-optically synthesize and project super-resolved images at its output FOV. Our results indicate that this diffractive image display can achieve a super-resolution factor of ~4, demonstrating a ~16-fold increase in SBP. We also experimentally validate the success of this diffractive super-resolution display using 3D-printed diffractive decoders that operate at the THz spectrum. This diffractive image decoder can be scaled to operate at visible wavelengths and inspire the design of large FOV and high-resolution displays that are compact, low-power, and computationally efficient.
ECG Heartbeat Classification Using Multimodal Fusion
Jul 21, 2021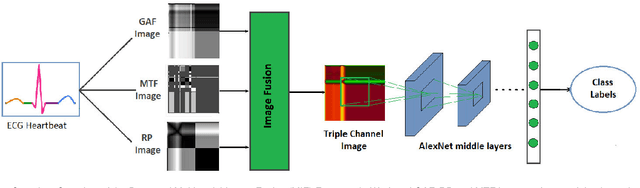
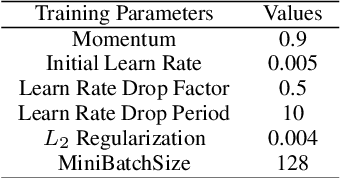
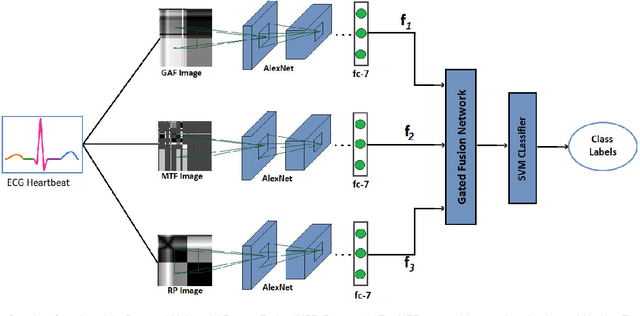
Abstract:Electrocardiogram (ECG) is an authoritative source to diagnose and counter critical cardiovascular syndromes such as arrhythmia and myocardial infarction (MI). Current machine learning techniques either depend on manually extracted features or large and complex deep learning networks which merely utilize the 1D ECG signal directly. Since intelligent multimodal fusion can perform at the stateof-the-art level with an efficient deep network, therefore, in this paper, we propose two computationally efficient multimodal fusion frameworks for ECG heart beat classification called Multimodal Image Fusion (MIF) and Multimodal Feature Fusion (MFF). At the input of these frameworks, we convert the raw ECG data into three different images using Gramian Angular Field (GAF), Recurrence Plot (RP) and Markov Transition Field (MTF). In MIF, we first perform image fusion by combining three imaging modalities to create a single image modality which serves as input to the Convolutional Neural Network (CNN). In MFF, we extracted features from penultimate layer of CNNs and fused them to get unique and interdependent information necessary for better performance of classifier. These informational features are finally used to train a Support Vector Machine (SVM) classifier for ECG heart-beat classification. We demonstrate the superiority of the proposed fusion models by performing experiments on PhysioNets MIT-BIH dataset for five distinct conditions of arrhythmias which are consistent with the AAMI EC57 protocols and on PTB diagnostics dataset for Myocardial Infarction (MI) classification. We achieved classification accuracy of 99.7% and 99.2% on arrhythmia and MI classification, respectively.
ECG Heart-beat Classification Using Multimodal Image Fusion
May 28, 2021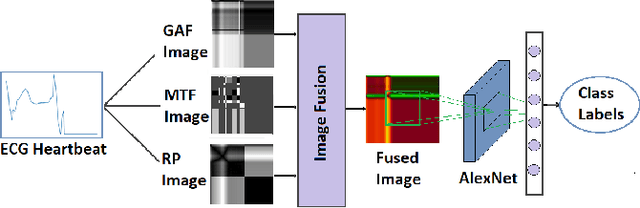
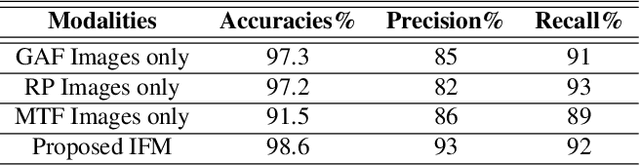
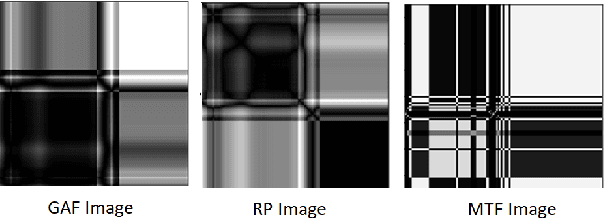
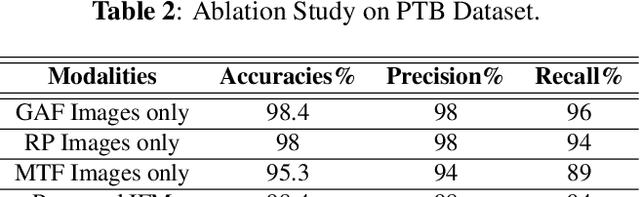
Abstract:In this paper, we present a novel Image Fusion Model (IFM) for ECG heart-beat classification to overcome the weaknesses of existing machine learning techniques that rely either on manual feature extraction or direct utilization of 1D raw ECG signal. At the input of IFM, we first convert the heart beats of ECG into three different images using Gramian Angular Field (GAF), Recurrence Plot (RP) and Markov Transition Field (MTF) and then fuse these images to create a single imaging modality. We use AlexNet for feature extraction and classification and thus employ end to end deep learning. We perform experiments on PhysioNet MIT-BIH dataset for five different arrhythmias in accordance with the AAMI EC57 standard and on PTB diagnostics dataset for myocardial infarction (MI) classification. We achieved an state of an art results in terms of prediction accuracy, precision and recall.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge