Aydogan Ozcan
Automated HER2 scoring with uncertainty quantification using lensfree holography and deep learning
Jan 26, 2026Abstract:Accurate assessment of human epidermal growth factor receptor 2 (HER2) expression is critical for breast cancer diagnosis, prognosis, and therapy selection; yet, most existing digital HER2 scoring methods rely on bulky and expensive optical systems. Here, we present a compact and cost-effective lensfree holography platform integrated with deep learning for automated HER2 scoring of immunohistochemically stained breast tissue sections. The system captures lensfree diffraction patterns of stained HER2 tissue sections under RGB laser illumination and acquires complex field information over a sample area of ~1,250 mm^2 at an effective throughput of ~84 mm^2 per minute. To enhance diagnostic reliability, we incorporated an uncertainty quantification strategy based on Bayesian Monte Carlo dropout, which provides autonomous uncertainty estimates for each prediction and supports reliable, robust HER2 scoring, with an overall correction rate of 30.4%. Using a blinded test set of 412 unique tissue samples, our approach achieved a testing accuracy of 84.9% for 4-class (0, 1+, 2+, 3+) HER2 classification and 94.8% for binary (0/1+ vs. 2+/3+) HER2 scoring with uncertainty quantification. Overall, this lensfree holography approach provides a practical pathway toward portable, high-throughput, and cost-effective HER2 scoring, particularly suited for resource-limited settings, where traditional digital pathology infrastructure is unavailable.
Autonomous Uncertainty Quantification for Computational Point-of-care Sensors
Dec 24, 2025Abstract:Computational point-of-care (POC) sensors enable rapid, low-cost, and accessible diagnostics in emergency, remote and resource-limited areas that lack access to centralized medical facilities. These systems can utilize neural network-based algorithms to accurately infer a diagnosis from the signals generated by rapid diagnostic tests or sensors. However, neural network-based diagnostic models are subject to hallucinations and can produce erroneous predictions, posing a risk of misdiagnosis and inaccurate clinical decisions. To address this challenge, here we present an autonomous uncertainty quantification technique developed for POC diagnostics. As our testbed, we used a paper-based, computational vertical flow assay (xVFA) platform developed for rapid POC diagnosis of Lyme disease, the most prevalent tick-borne disease globally. The xVFA platform integrates a disposable paper-based assay, a handheld optical reader and a neural network-based inference algorithm, providing rapid and cost-effective Lyme disease diagnostics in under 20 min using only 20 uL of patient serum. By incorporating a Monte Carlo dropout (MCDO)-based uncertainty quantification approach into the diagnostics pipeline, we identified and excluded erroneous predictions with high uncertainty, significantly improving the sensitivity and reliability of the xVFA in an autonomous manner, without access to the ground truth diagnostic information of patients. Blinded testing using new patient samples demonstrated an increase in diagnostic sensitivity from 88.2% to 95.7%, indicating the effectiveness of MCDO-based uncertainty quantification in enhancing the robustness of neural network-driven computational POC sensing systems.
Deep learning-enhanced dual-mode multiplexed optical sensor for point-of-care diagnostics of cardiovascular diseases
Dec 24, 2025Abstract:Rapid and accessible cardiac biomarker testing is essential for the timely diagnosis and risk assessment of myocardial infarction (MI) and heart failure (HF), two interrelated conditions that frequently coexist and drive recurrent hospitalizations with high mortality. However, current laboratory and point-of-care testing systems are limited by long turnaround times, narrow dynamic ranges for the tested biomarkers, and single-analyte formats that fail to capture the complexity of cardiovascular disease. Here, we present a deep learning-enhanced dual-mode multiplexed vertical flow assay (xVFA) with a portable optical reader and a neural network-based quantification pipeline. This optical sensor integrates colorimetric and chemiluminescent detection within a single paper-based cartridge to complementarily cover a large dynamic range (spanning ~6 orders of magnitude) for both low- and high-abundance biomarkers, while maintaining quantitative accuracy. Using 50 uL of serum, the optical sensor simultaneously quantifies cardiac troponin I (cTnI), creatine kinase-MB (CK-MB), and N-terminal pro-B-type natriuretic peptide (NT-proBNP) within 23 min. The xVFA achieves sub-pg/mL sensitivity for cTnI and sub-ng/mL sensitivity for CK-MB and NT-proBNP, spanning the clinically relevant ranges for these biomarkers. Neural network models trained and blindly tested on 92 patient serum samples yielded a robust quantification performance (Pearson's r > 0.96 vs. reference assays). By combining high sensitivity, multiplexing, and automation in a compact and cost-effective optical sensor format, the dual-mode xVFA enables rapid and quantitative cardiovascular diagnostics at the point of care.
Snapshot 3D image projection using a diffractive decoder
Dec 23, 2025Abstract:3D image display is essential for next-generation volumetric imaging; however, dense depth multiplexing for 3D image projection remains challenging because diffraction-induced cross-talk rapidly increases as the axial image planes get closer. Here, we introduce a 3D display system comprising a digital encoder and a diffractive optical decoder, which simultaneously projects different images onto multiple target axial planes with high axial resolution. By leveraging multi-layer diffractive wavefront decoding and deep learning-based end-to-end optimization, the system achieves high-fidelity depth-resolved 3D image projection in a snapshot, enabling axial plane separations on the order of a wavelength. The digital encoder leverages a Fourier encoder network to capture multi-scale spatial and frequency-domain features from input images, integrates axial position encoding, and generates a unified phase representation that simultaneously encodes all images to be axially projected in a single snapshot through a jointly-optimized diffractive decoder. We characterized the impact of diffractive decoder depth, output diffraction efficiency, spatial light modulator resolution, and axial encoding density, revealing trade-offs that govern axial separation and 3D image projection quality. We further demonstrated the capability to display volumetric images containing 28 axial slices, as well as the ability to dynamically reconfigure the axial locations of the image planes, performed on demand. Finally, we experimentally validated the presented approach, demonstrating close agreement between the measured results and the target images. These results establish the diffractive 3D display system as a compact and scalable framework for depth-resolved snapshot 3D image projection, with potential applications in holographic displays, AR/VR interfaces, and volumetric optical computing.
Pixel Super-Resolved Fluorescence Lifetime Imaging Using Deep Learning
Dec 18, 2025



Abstract:Fluorescence lifetime imaging microscopy (FLIM) is a powerful quantitative technique that provides metabolic and molecular contrast, offering strong translational potential for label-free, real-time diagnostics. However, its clinical adoption remains limited by long pixel dwell times and low signal-to-noise ratio (SNR), which impose a stricter resolution-speed trade-off than conventional optical imaging approaches. Here, we introduce FLIM_PSR_k, a deep learning-based multi-channel pixel super-resolution (PSR) framework that reconstructs high-resolution FLIM images from data acquired with up to a 5-fold increased pixel size. The model is trained using the conditional generative adversarial network (cGAN) framework, which, compared to diffusion model-based alternatives, delivers a more robust PSR reconstruction with substantially shorter inference times, a crucial advantage for practical deployment. FLIM_PSR_k not only enables faster image acquisition but can also alleviate SNR limitations in autofluorescence-based FLIM. Blind testing on held-out patient-derived tumor tissue samples demonstrates that FLIM_PSR_k reliably achieves a super-resolution factor of k = 5, resulting in a 25-fold increase in the space-bandwidth product of the output images and revealing fine architectural features lost in lower-resolution inputs, with statistically significant improvements across various image quality metrics. By increasing FLIM's effective spatial resolution, FLIM_PSR_k advances lifetime imaging toward faster, higher-resolution, and hardware-flexible implementations compatible with low-numerical-aperture and miniaturized platforms, better positioning FLIM for translational applications.
Universal point spread function engineering for 3D optical information processing
Feb 09, 2025Abstract:Point spread function (PSF) engineering has been pivotal in the remarkable progress made in high-resolution imaging in the last decades. However, the diversity in PSF structures attainable through existing engineering methods is limited. Here, we report universal PSF engineering, demonstrating a method to synthesize an arbitrary set of spatially varying 3D PSFs between the input and output volumes of a spatially incoherent diffractive processor composed of cascaded transmissive surfaces. We rigorously analyze the PSF engineering capabilities of such diffractive processors within the diffraction limit of light and provide numerical demonstrations of unique imaging capabilities, such as snapshot 3D multispectral imaging without involving any spectral filters, axial scanning or digital reconstruction steps, which is enabled by the spatial and spectral engineering of 3D PSFs. Our framework and analysis would be important for future advancements in computational imaging, sensing and diffractive processing of 3D optical information.
Snapshot multi-spectral imaging through defocusing and a Fourier imager network
Jan 24, 2025



Abstract:Multi-spectral imaging, which simultaneously captures the spatial and spectral information of a scene, is widely used across diverse fields, including remote sensing, biomedical imaging, and agricultural monitoring. Here, we introduce a snapshot multi-spectral imaging approach employing a standard monochrome image sensor with no additional spectral filters or customized components. Our system leverages the inherent chromatic aberration of wavelength-dependent defocusing as a natural source of physical encoding of multi-spectral information; this encoded image information is rapidly decoded via a deep learning-based multi-spectral Fourier Imager Network (mFIN). We experimentally tested our method with six illumination bands and demonstrated an overall accuracy of 92.98% for predicting the illumination channels at the input and achieved a robust multi-spectral image reconstruction on various test objects. This deep learning-powered framework achieves high-quality multi-spectral image reconstruction using snapshot image acquisition with a monochrome image sensor and could be useful for applications in biomedicine, industrial quality control, and agriculture, among others.
Virtual Staining of Label-Free Tissue in Imaging Mass Spectrometry
Nov 20, 2024



Abstract:Imaging mass spectrometry (IMS) is a powerful tool for untargeted, highly multiplexed molecular mapping of tissue in biomedical research. IMS offers a means of mapping the spatial distributions of molecular species in biological tissue with unparalleled chemical specificity and sensitivity. However, most IMS platforms are not able to achieve microscopy-level spatial resolution and lack cellular morphological contrast, necessitating subsequent histochemical staining, microscopic imaging and advanced image registration steps to enable molecular distributions to be linked to specific tissue features and cell types. Here, we present a virtual histological staining approach that enhances spatial resolution and digitally introduces cellular morphological contrast into mass spectrometry images of label-free human tissue using a diffusion model. Blind testing on human kidney tissue demonstrated that the virtually stained images of label-free samples closely match their histochemically stained counterparts (with Periodic Acid-Schiff staining), showing high concordance in identifying key renal pathology structures despite utilizing IMS data with 10-fold larger pixel size. Additionally, our approach employs an optimized noise sampling technique during the diffusion model's inference process to reduce variance in the generated images, yielding reliable and repeatable virtual staining. We believe this virtual staining method will significantly expand the applicability of IMS in life sciences and open new avenues for mass spectrometry-based biomedical research.
Leveraging Computational Pathology AI for Noninvasive Optical Imaging Analysis Without Retraining
Nov 19, 2024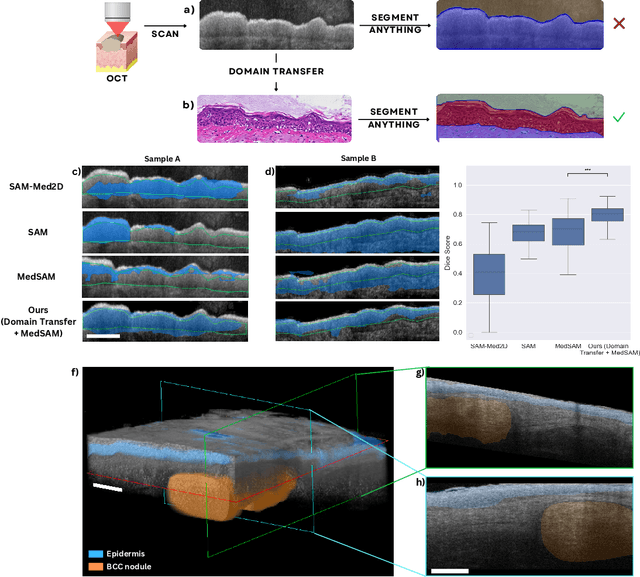
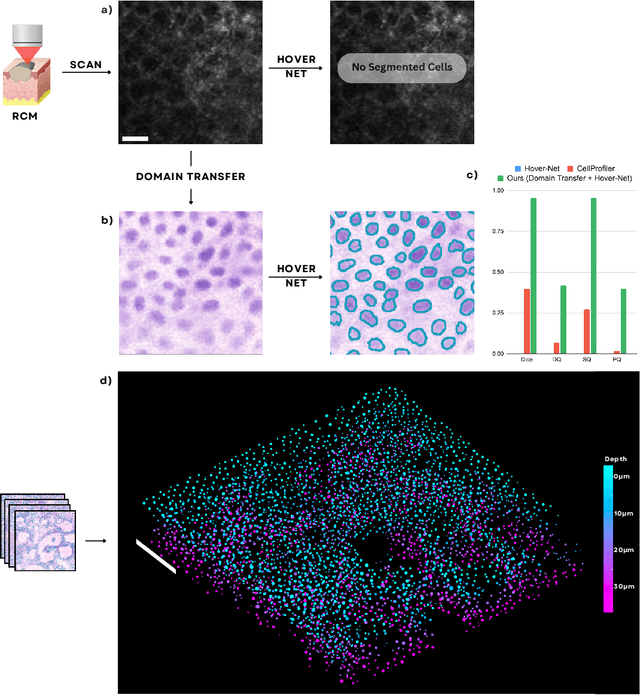
Abstract:Noninvasive optical imaging modalities can probe patient's tissue in 3D and over time generate gigabytes of clinically relevant data per sample. There is a need for AI models to analyze this data and assist clinical workflow. The lack of expert labelers and the large dataset required (>100,000 images) for model training and tuning are the main hurdles in creating foundation models. In this paper we introduce FoundationShift, a method to apply any AI model from computational pathology without retraining. We show our method is more accurate than state of the art models (SAM, MedSAM, SAM-Med2D, CellProfiler, Hover-Net, PLIP, UNI and ChatGPT), with multiple imaging modalities (OCT and RCM). This is achieved without the need for model retraining or fine-tuning. Applying our method to noninvasive in vivo images could enable physicians to readily incorporate optical imaging modalities into their clinical practice, providing real time tissue analysis and improving patient care.
Super-resolved virtual staining of label-free tissue using diffusion models
Oct 26, 2024



Abstract:Virtual staining of tissue offers a powerful tool for transforming label-free microscopy images of unstained tissue into equivalents of histochemically stained samples. This study presents a diffusion model-based super-resolution virtual staining approach utilizing a Brownian bridge process to enhance both the spatial resolution and fidelity of label-free virtual tissue staining, addressing the limitations of traditional deep learning-based methods. Our approach integrates novel sampling techniques into a diffusion model-based image inference process to significantly reduce the variance in the generated virtually stained images, resulting in more stable and accurate outputs. Blindly applied to lower-resolution auto-fluorescence images of label-free human lung tissue samples, the diffusion-based super-resolution virtual staining model consistently outperformed conventional approaches in resolution, structural similarity and perceptual accuracy, successfully achieving a super-resolution factor of 4-5x, increasing the output space-bandwidth product by 16-25-fold compared to the input label-free microscopy images. Diffusion-based super-resolved virtual tissue staining not only improves resolution and image quality but also enhances the reliability of virtual staining without traditional chemical staining, offering significant potential for clinical diagnostics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge