Luzhe Huang
Snapshot multi-spectral imaging through defocusing and a Fourier imager network
Jan 24, 2025



Abstract:Multi-spectral imaging, which simultaneously captures the spatial and spectral information of a scene, is widely used across diverse fields, including remote sensing, biomedical imaging, and agricultural monitoring. Here, we introduce a snapshot multi-spectral imaging approach employing a standard monochrome image sensor with no additional spectral filters or customized components. Our system leverages the inherent chromatic aberration of wavelength-dependent defocusing as a natural source of physical encoding of multi-spectral information; this encoded image information is rapidly decoded via a deep learning-based multi-spectral Fourier Imager Network (mFIN). We experimentally tested our method with six illumination bands and demonstrated an overall accuracy of 92.98% for predicting the illumination channels at the input and achieved a robust multi-spectral image reconstruction on various test objects. This deep learning-powered framework achieves high-quality multi-spectral image reconstruction using snapshot image acquisition with a monochrome image sensor and could be useful for applications in biomedicine, industrial quality control, and agriculture, among others.
Virtual Staining of Label-Free Tissue in Imaging Mass Spectrometry
Nov 20, 2024



Abstract:Imaging mass spectrometry (IMS) is a powerful tool for untargeted, highly multiplexed molecular mapping of tissue in biomedical research. IMS offers a means of mapping the spatial distributions of molecular species in biological tissue with unparalleled chemical specificity and sensitivity. However, most IMS platforms are not able to achieve microscopy-level spatial resolution and lack cellular morphological contrast, necessitating subsequent histochemical staining, microscopic imaging and advanced image registration steps to enable molecular distributions to be linked to specific tissue features and cell types. Here, we present a virtual histological staining approach that enhances spatial resolution and digitally introduces cellular morphological contrast into mass spectrometry images of label-free human tissue using a diffusion model. Blind testing on human kidney tissue demonstrated that the virtually stained images of label-free samples closely match their histochemically stained counterparts (with Periodic Acid-Schiff staining), showing high concordance in identifying key renal pathology structures despite utilizing IMS data with 10-fold larger pixel size. Additionally, our approach employs an optimized noise sampling technique during the diffusion model's inference process to reduce variance in the generated images, yielding reliable and repeatable virtual staining. We believe this virtual staining method will significantly expand the applicability of IMS in life sciences and open new avenues for mass spectrometry-based biomedical research.
Multi-scale Generative Modeling for Fast Sampling
Nov 14, 2024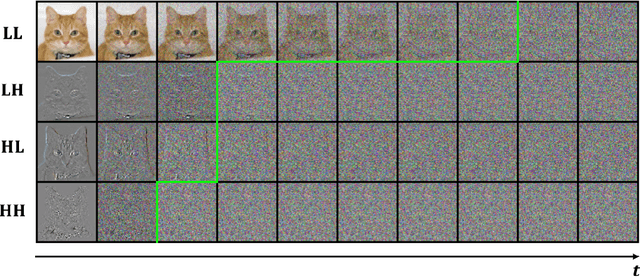

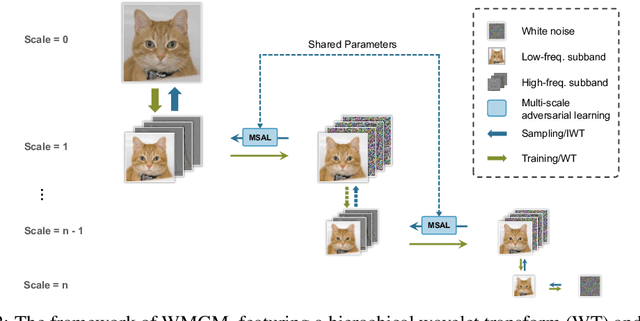
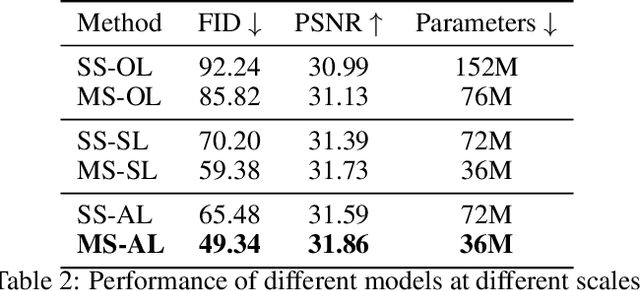
Abstract:While working within the spatial domain can pose problems associated with ill-conditioned scores caused by power-law decay, recent advances in diffusion-based generative models have shown that transitioning to the wavelet domain offers a promising alternative. However, within the wavelet domain, we encounter unique challenges, especially the sparse representation of high-frequency coefficients, which deviates significantly from the Gaussian assumptions in the diffusion process. To this end, we propose a multi-scale generative modeling in the wavelet domain that employs distinct strategies for handling low and high-frequency bands. In the wavelet domain, we apply score-based generative modeling with well-conditioned scores for low-frequency bands, while utilizing a multi-scale generative adversarial learning for high-frequency bands. As supported by the theoretical analysis and experimental results, our model significantly improve performance and reduce the number of trainable parameters, sampling steps, and time.
Super-resolved virtual staining of label-free tissue using diffusion models
Oct 26, 2024



Abstract:Virtual staining of tissue offers a powerful tool for transforming label-free microscopy images of unstained tissue into equivalents of histochemically stained samples. This study presents a diffusion model-based super-resolution virtual staining approach utilizing a Brownian bridge process to enhance both the spatial resolution and fidelity of label-free virtual tissue staining, addressing the limitations of traditional deep learning-based methods. Our approach integrates novel sampling techniques into a diffusion model-based image inference process to significantly reduce the variance in the generated virtually stained images, resulting in more stable and accurate outputs. Blindly applied to lower-resolution auto-fluorescence images of label-free human lung tissue samples, the diffusion-based super-resolution virtual staining model consistently outperformed conventional approaches in resolution, structural similarity and perceptual accuracy, successfully achieving a super-resolution factor of 4-5x, increasing the output space-bandwidth product by 16-25-fold compared to the input label-free microscopy images. Diffusion-based super-resolved virtual tissue staining not only improves resolution and image quality but also enhances the reliability of virtual staining without traditional chemical staining, offering significant potential for clinical diagnostics.
Unidirectional imaging with partially coherent light
Aug 10, 2024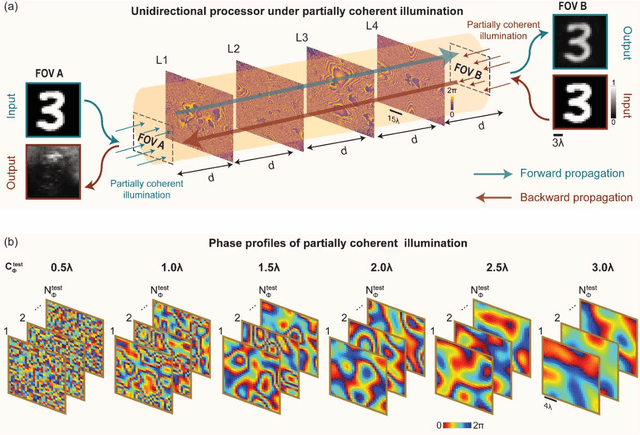
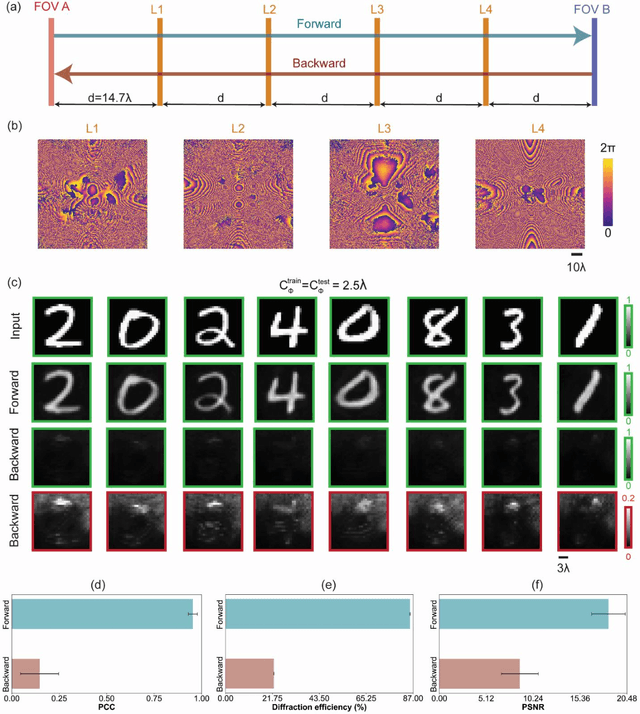
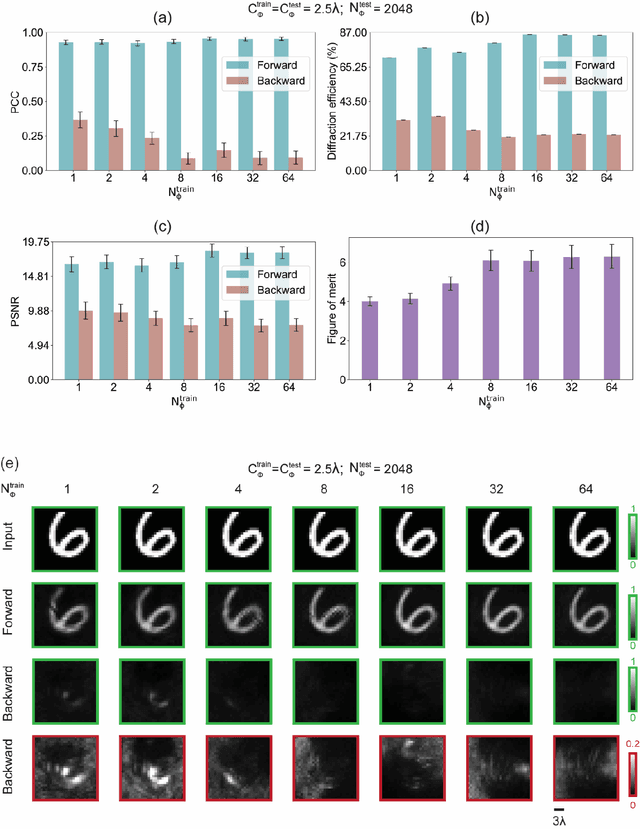

Abstract:Unidirectional imagers form images of input objects only in one direction, e.g., from field-of-view (FOV) A to FOV B, while blocking the image formation in the reverse direction, from FOV B to FOV A. Here, we report unidirectional imaging under spatially partially coherent light and demonstrate high-quality imaging only in the forward direction (A->B) with high power efficiency while distorting the image formation in the backward direction (B->A) along with low power efficiency. Our reciprocal design features a set of spatially engineered linear diffractive layers that are statistically optimized for partially coherent illumination with a given phase correlation length. Our analyses reveal that when illuminated by a partially coherent beam with a correlation length of ~1.5 w or larger, where w is the wavelength of light, diffractive unidirectional imagers achieve robust performance, exhibiting asymmetric imaging performance between the forward and backward directions - as desired. A partially coherent unidirectional imager designed with a smaller correlation length of less than 1.5 w still supports unidirectional image transmission, but with a reduced figure of merit. These partially coherent diffractive unidirectional imagers are compact (axially spanning less than 75 w), polarization-independent, and compatible with various types of illumination sources, making them well-suited for applications in asymmetric visual information processing and communication.
Multi-scale Conditional Generative Modeling for Microscopic Image Restoration
Jul 07, 2024



Abstract:The advance of diffusion-based generative models in recent years has revolutionized state-of-the-art (SOTA) techniques in a wide variety of image analysis and synthesis tasks, whereas their adaptation on image restoration, particularly within computational microscopy remains theoretically and empirically underexplored. In this research, we introduce a multi-scale generative model that enhances conditional image restoration through a novel exploitation of the Brownian Bridge process within wavelet domain. By initiating the Brownian Bridge diffusion process specifically at the lowest-frequency subband and applying generative adversarial networks at subsequent multi-scale high-frequency subbands in the wavelet domain, our method provides significant acceleration during training and sampling while sustaining a high image generation quality and diversity on par with SOTA diffusion models. Experimental results on various computational microscopy and imaging tasks confirm our method's robust performance and its considerable reduction in its sampling steps and time. This pioneering technique offers an efficient image restoration framework that harmonizes efficiency with quality, signifying a major stride in incorporating cutting-edge generative models into computational microscopy workflows.
Autonomous Quality and Hallucination Assessment for Virtual Tissue Staining and Digital Pathology
Apr 29, 2024Abstract:Histopathological staining of human tissue is essential in the diagnosis of various diseases. The recent advances in virtual tissue staining technologies using AI alleviate some of the costly and tedious steps involved in the traditional histochemical staining process, permitting multiplexed rapid staining of label-free tissue without using staining reagents, while also preserving tissue. However, potential hallucinations and artifacts in these virtually stained tissue images pose concerns, especially for the clinical utility of these approaches. Quality assessment of histology images is generally performed by human experts, which can be subjective and depends on the training level of the expert. Here, we present an autonomous quality and hallucination assessment method (termed AQuA), mainly designed for virtual tissue staining, while also being applicable to histochemical staining. AQuA achieves 99.8% accuracy when detecting acceptable and unacceptable virtually stained tissue images without access to ground truth, also presenting an agreement of 98.5% with the manual assessments made by board-certified pathologists. Besides, AQuA achieves super-human performance in identifying realistic-looking, virtually stained hallucinatory images that would normally mislead human diagnosticians by deceiving them into diagnosing patients that never existed. We further demonstrate the wide adaptability of AQuA across various virtually and histochemically stained tissue images and showcase its strong external generalization to detect unseen hallucination patterns of virtual staining network models as well as artifacts observed in the traditional histochemical staining workflow. This framework creates new opportunities to enhance the reliability of virtual staining and will provide quality assurance for various image generation and transformation tasks in digital pathology and computational imaging.
Neural Network-Based Processing and Reconstruction of Compromised Biophotonic Image Data
Mar 21, 2024Abstract:The integration of deep learning techniques with biophotonic setups has opened new horizons in bioimaging. A compelling trend in this field involves deliberately compromising certain measurement metrics to engineer better bioimaging tools in terms of cost, speed, and form-factor, followed by compensating for the resulting defects through the utilization of deep learning models trained on a large amount of ideal, superior or alternative data. This strategic approach has found increasing popularity due to its potential to enhance various aspects of biophotonic imaging. One of the primary motivations for employing this strategy is the pursuit of higher temporal resolution or increased imaging speed, critical for capturing fine dynamic biological processes. This approach also offers the prospect of simplifying hardware requirements/complexities, thereby making advanced imaging standards more accessible in terms of cost and/or size. This article provides an in-depth review of the diverse measurement aspects that researchers intentionally impair in their biophotonic setups, including the point spread function, signal-to-noise ratio, sampling density, and pixel resolution. By deliberately compromising these metrics, researchers aim to not only recuperate them through the application of deep learning networks, but also bolster in return other crucial parameters, such as the field-of-view, depth-of-field, and space-bandwidth product. Here, we discuss various biophotonic methods that have successfully employed this strategic approach. These techniques span broad applications and showcase the versatility and effectiveness of deep learning in the context of compromised biophotonic data. Finally, by offering our perspectives on the future possibilities of this rapidly evolving concept, we hope to motivate our readers to explore novel ways of balancing hardware compromises with compensation via AI.
Virtual histological staining of unlabeled autopsy tissue
Aug 02, 2023Abstract:Histological examination is a crucial step in an autopsy; however, the traditional histochemical staining of post-mortem samples faces multiple challenges, including the inferior staining quality due to autolysis caused by delayed fixation of cadaver tissue, as well as the resource-intensive nature of chemical staining procedures covering large tissue areas, which demand substantial labor, cost, and time. These challenges can become more pronounced during global health crises when the availability of histopathology services is limited, resulting in further delays in tissue fixation and more severe staining artifacts. Here, we report the first demonstration of virtual staining of autopsy tissue and show that a trained neural network can rapidly transform autofluorescence images of label-free autopsy tissue sections into brightfield equivalent images that match hematoxylin and eosin (H&E) stained versions of the same samples, eliminating autolysis-induced severe staining artifacts inherent in traditional histochemical staining of autopsied tissue. Our virtual H&E model was trained using >0.7 TB of image data and a data-efficient collaboration scheme that integrates the virtual staining network with an image registration network. The trained model effectively accentuated nuclear, cytoplasmic and extracellular features in new autopsy tissue samples that experienced severe autolysis, such as COVID-19 samples never seen before, where the traditional histochemical staining failed to provide consistent staining quality. This virtual autopsy staining technique can also be extended to necrotic tissue, and can rapidly and cost-effectively generate artifact-free H&E stains despite severe autolysis and cell death, also reducing labor, cost and infrastructure requirements associated with the standard histochemical staining.
Cycle Consistency-based Uncertainty Quantification of Neural Networks in Inverse Imaging Problems
May 22, 2023Abstract:Uncertainty estimation is critical for numerous applications of deep neural networks and draws growing attention from researchers. Here, we demonstrate an uncertainty quantification approach for deep neural networks used in inverse problems based on cycle consistency. We build forward-backward cycles using the physical forward model available and a trained deep neural network solving the inverse problem at hand, and accordingly derive uncertainty estimators through regression analysis on the consistency of these forward-backward cycles. We theoretically analyze cycle consistency metrics and derive their relationship with respect to uncertainty, bias, and robustness of the neural network inference. To demonstrate the effectiveness of these cycle consistency-based uncertainty estimators, we classified corrupted and out-of-distribution input image data using some of the widely used image deblurring and super-resolution neural networks as testbeds. The blind testing of our method outperformed other models in identifying unseen input data corruption and distribution shifts. This work provides a simple-to-implement and rapid uncertainty quantification method that can be universally applied to various neural networks used for solving inverse problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge