Yuzhu Li
Automated HER2 scoring with uncertainty quantification using lensfree holography and deep learning
Jan 26, 2026Abstract:Accurate assessment of human epidermal growth factor receptor 2 (HER2) expression is critical for breast cancer diagnosis, prognosis, and therapy selection; yet, most existing digital HER2 scoring methods rely on bulky and expensive optical systems. Here, we present a compact and cost-effective lensfree holography platform integrated with deep learning for automated HER2 scoring of immunohistochemically stained breast tissue sections. The system captures lensfree diffraction patterns of stained HER2 tissue sections under RGB laser illumination and acquires complex field information over a sample area of ~1,250 mm^2 at an effective throughput of ~84 mm^2 per minute. To enhance diagnostic reliability, we incorporated an uncertainty quantification strategy based on Bayesian Monte Carlo dropout, which provides autonomous uncertainty estimates for each prediction and supports reliable, robust HER2 scoring, with an overall correction rate of 30.4%. Using a blinded test set of 412 unique tissue samples, our approach achieved a testing accuracy of 84.9% for 4-class (0, 1+, 2+, 3+) HER2 classification and 94.8% for binary (0/1+ vs. 2+/3+) HER2 scoring with uncertainty quantification. Overall, this lensfree holography approach provides a practical pathway toward portable, high-throughput, and cost-effective HER2 scoring, particularly suited for resource-limited settings, where traditional digital pathology infrastructure is unavailable.
Deep learning-enhanced dual-mode multiplexed optical sensor for point-of-care diagnostics of cardiovascular diseases
Dec 24, 2025Abstract:Rapid and accessible cardiac biomarker testing is essential for the timely diagnosis and risk assessment of myocardial infarction (MI) and heart failure (HF), two interrelated conditions that frequently coexist and drive recurrent hospitalizations with high mortality. However, current laboratory and point-of-care testing systems are limited by long turnaround times, narrow dynamic ranges for the tested biomarkers, and single-analyte formats that fail to capture the complexity of cardiovascular disease. Here, we present a deep learning-enhanced dual-mode multiplexed vertical flow assay (xVFA) with a portable optical reader and a neural network-based quantification pipeline. This optical sensor integrates colorimetric and chemiluminescent detection within a single paper-based cartridge to complementarily cover a large dynamic range (spanning ~6 orders of magnitude) for both low- and high-abundance biomarkers, while maintaining quantitative accuracy. Using 50 uL of serum, the optical sensor simultaneously quantifies cardiac troponin I (cTnI), creatine kinase-MB (CK-MB), and N-terminal pro-B-type natriuretic peptide (NT-proBNP) within 23 min. The xVFA achieves sub-pg/mL sensitivity for cTnI and sub-ng/mL sensitivity for CK-MB and NT-proBNP, spanning the clinically relevant ranges for these biomarkers. Neural network models trained and blindly tested on 92 patient serum samples yielded a robust quantification performance (Pearson's r > 0.96 vs. reference assays). By combining high sensitivity, multiplexing, and automation in a compact and cost-effective optical sensor format, the dual-mode xVFA enables rapid and quantitative cardiovascular diagnostics at the point of care.
Virtual Staining of Label-Free Tissue in Imaging Mass Spectrometry
Nov 20, 2024



Abstract:Imaging mass spectrometry (IMS) is a powerful tool for untargeted, highly multiplexed molecular mapping of tissue in biomedical research. IMS offers a means of mapping the spatial distributions of molecular species in biological tissue with unparalleled chemical specificity and sensitivity. However, most IMS platforms are not able to achieve microscopy-level spatial resolution and lack cellular morphological contrast, necessitating subsequent histochemical staining, microscopic imaging and advanced image registration steps to enable molecular distributions to be linked to specific tissue features and cell types. Here, we present a virtual histological staining approach that enhances spatial resolution and digitally introduces cellular morphological contrast into mass spectrometry images of label-free human tissue using a diffusion model. Blind testing on human kidney tissue demonstrated that the virtually stained images of label-free samples closely match their histochemically stained counterparts (with Periodic Acid-Schiff staining), showing high concordance in identifying key renal pathology structures despite utilizing IMS data with 10-fold larger pixel size. Additionally, our approach employs an optimized noise sampling technique during the diffusion model's inference process to reduce variance in the generated images, yielding reliable and repeatable virtual staining. We believe this virtual staining method will significantly expand the applicability of IMS in life sciences and open new avenues for mass spectrometry-based biomedical research.
Super-resolved virtual staining of label-free tissue using diffusion models
Oct 26, 2024



Abstract:Virtual staining of tissue offers a powerful tool for transforming label-free microscopy images of unstained tissue into equivalents of histochemically stained samples. This study presents a diffusion model-based super-resolution virtual staining approach utilizing a Brownian bridge process to enhance both the spatial resolution and fidelity of label-free virtual tissue staining, addressing the limitations of traditional deep learning-based methods. Our approach integrates novel sampling techniques into a diffusion model-based image inference process to significantly reduce the variance in the generated virtually stained images, resulting in more stable and accurate outputs. Blindly applied to lower-resolution auto-fluorescence images of label-free human lung tissue samples, the diffusion-based super-resolution virtual staining model consistently outperformed conventional approaches in resolution, structural similarity and perceptual accuracy, successfully achieving a super-resolution factor of 4-5x, increasing the output space-bandwidth product by 16-25-fold compared to the input label-free microscopy images. Diffusion-based super-resolved virtual tissue staining not only improves resolution and image quality but also enhances the reliability of virtual staining without traditional chemical staining, offering significant potential for clinical diagnostics.
Deep Learning-based Detection of Bacterial Swarm Motion Using a Single Image
Oct 19, 2024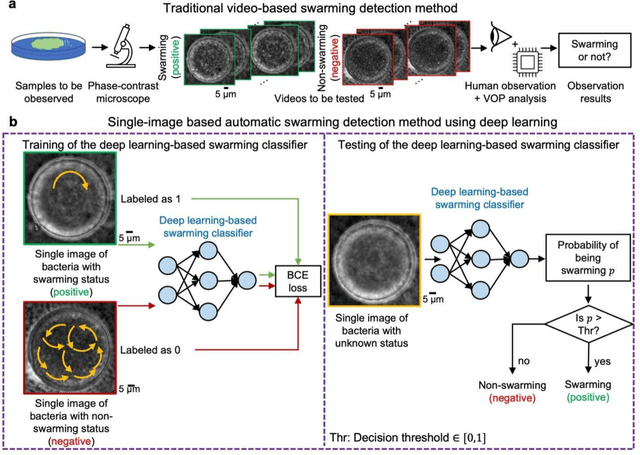
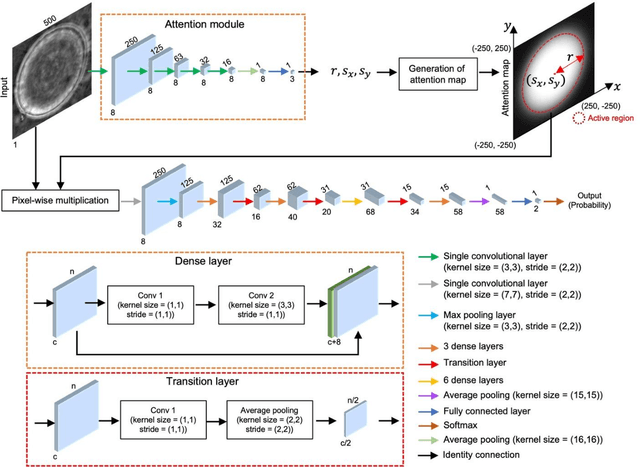
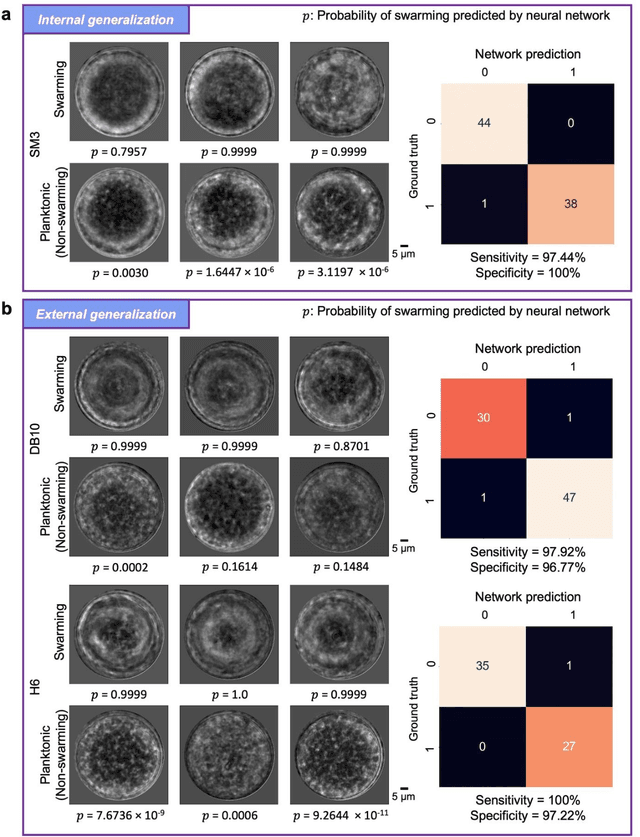
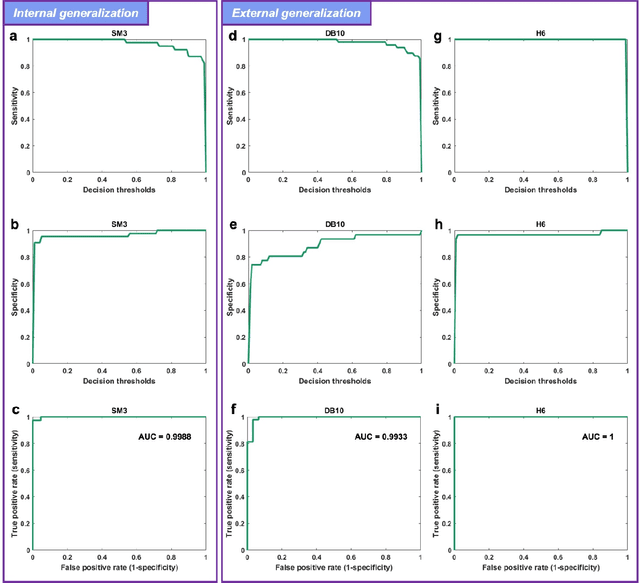
Abstract:Distinguishing between swarming and swimming, the two principal forms of bacterial movement, holds significant conceptual and clinical relevance. This is because bacteria that exhibit swarming capabilities often possess unique properties crucial to the pathogenesis of infectious diseases and may also have therapeutic potential. Here, we report a deep learning-based swarming classifier that rapidly and autonomously predicts swarming probability using a single blurry image. Compared with traditional video-based, manually-processed approaches, our method is particularly suited for high-throughput environments and provides objective, quantitative assessments of swarming probability. The swarming classifier demonstrated in our work was trained on Enterobacter sp. SM3 and showed good performance when blindly tested on new swarming (positive) and swimming (negative) test images of SM3, achieving a sensitivity of 97.44% and a specificity of 100%. Furthermore, this classifier demonstrated robust external generalization capabilities when applied to unseen bacterial species, such as Serratia marcescens DB10 and Citrobacter koseri H6. It blindly achieved a sensitivity of 97.92% and a specificity of 96.77% for DB10, and a sensitivity of 100% and a specificity of 97.22% for H6. This competitive performance indicates the potential to adapt our approach for diagnostic applications through portable devices or even smartphones. This adaptation would facilitate rapid, objective, on-site screening for bacterial swarming motility, potentially enhancing the early detection and treatment assessment of various diseases, including inflammatory bowel diseases (IBD) and urinary tract infections (UTI).
Label-free evaluation of lung and heart transplant biopsies using virtual staining
Sep 09, 2024



Abstract:Organ transplantation serves as the primary therapeutic strategy for end-stage organ failures. However, allograft rejection is a common complication of organ transplantation. Histological assessment is essential for the timely detection and diagnosis of transplant rejection and remains the gold standard. Nevertheless, the traditional histochemical staining process is time-consuming, costly, and labor-intensive. Here, we present a panel of virtual staining neural networks for lung and heart transplant biopsies, which digitally convert autofluorescence microscopic images of label-free tissue sections into their brightfield histologically stained counterparts, bypassing the traditional histochemical staining process. Specifically, we virtually generated Hematoxylin and Eosin (H&E), Masson's Trichrome (MT), and Elastic Verhoeff-Van Gieson (EVG) stains for label-free transplant lung tissue, along with H&E and MT stains for label-free transplant heart tissue. Subsequent blind evaluations conducted by three board-certified pathologists have confirmed that the virtual staining networks consistently produce high-quality histology images with high color uniformity, closely resembling their well-stained histochemical counterparts across various tissue features. The use of virtually stained images for the evaluation of transplant biopsies achieved comparable diagnostic outcomes to those obtained via traditional histochemical staining, with a concordance rate of 82.4% for lung samples and 91.7% for heart samples. Moreover, virtual staining models create multiple stains from the same autofluorescence input, eliminating structural mismatches observed between adjacent sections stained in the traditional workflow, while also saving tissue, expert time, and staining costs.
A Swift and Omnidirectional Formation Approach based on Hierarchical Reorganization
Jun 17, 2024Abstract:Current formations commonly rely on invariant hierarchical structures, such as predetermined leaders or enumerated formation shapes. These structures could be unidirectional and sluggish, constraining their adaptability and agility when encountering cluttered environments. To surmount these constraints, this work proposes an omnidirectional affine formation approach with hierarchical reorganizations. We first delineate the critical conditions requisite for facilitating hierarchical reorganizations within formations, which informs the development of the omnidirectional affine criterion. Central to our approach is the fluid leadership and authority redistribution, for which we develop a minimum time-driven leadership evaluation algorithm and a power transition control algorithm. These algorithms facilitate autonomous leader selection and ensure smooth power transitions, enabling the swarm to adapt hierarchically in alignment with the external environment. Furthermore, we deploy a power-centric topology switching mechanism tailored for the dynamic reorganization of in-team connections. Finally, simulations and experiments demonstrate the performance of the proposed method. The formation successfully performs several hierarchical reorganizations, with the longest reorganization taking only 0.047s. This swift adaptability allows five aerial robots to carry out complex tasks, including executing swerving movements and navigating through hoops at velocities up to 1.9m/s.
Autonomous Quality and Hallucination Assessment for Virtual Tissue Staining and Digital Pathology
Apr 29, 2024Abstract:Histopathological staining of human tissue is essential in the diagnosis of various diseases. The recent advances in virtual tissue staining technologies using AI alleviate some of the costly and tedious steps involved in the traditional histochemical staining process, permitting multiplexed rapid staining of label-free tissue without using staining reagents, while also preserving tissue. However, potential hallucinations and artifacts in these virtually stained tissue images pose concerns, especially for the clinical utility of these approaches. Quality assessment of histology images is generally performed by human experts, which can be subjective and depends on the training level of the expert. Here, we present an autonomous quality and hallucination assessment method (termed AQuA), mainly designed for virtual tissue staining, while also being applicable to histochemical staining. AQuA achieves 99.8% accuracy when detecting acceptable and unacceptable virtually stained tissue images without access to ground truth, also presenting an agreement of 98.5% with the manual assessments made by board-certified pathologists. Besides, AQuA achieves super-human performance in identifying realistic-looking, virtually stained hallucinatory images that would normally mislead human diagnosticians by deceiving them into diagnosing patients that never existed. We further demonstrate the wide adaptability of AQuA across various virtually and histochemically stained tissue images and showcase its strong external generalization to detect unseen hallucination patterns of virtual staining network models as well as artifacts observed in the traditional histochemical staining workflow. This framework creates new opportunities to enhance the reliability of virtual staining and will provide quality assurance for various image generation and transformation tasks in digital pathology and computational imaging.
Automated HER2 Scoring in Breast Cancer Images Using Deep Learning and Pyramid Sampling
Apr 01, 2024Abstract:Human epidermal growth factor receptor 2 (HER2) is a critical protein in cancer cell growth that signifies the aggressiveness of breast cancer (BC) and helps predict its prognosis. Accurate assessment of immunohistochemically (IHC) stained tissue slides for HER2 expression levels is essential for both treatment guidance and understanding of cancer mechanisms. Nevertheless, the traditional workflow of manual examination by board-certified pathologists encounters challenges, including inter- and intra-observer inconsistency and extended turnaround times. Here, we introduce a deep learning-based approach utilizing pyramid sampling for the automated classification of HER2 status in IHC-stained BC tissue images. Our approach analyzes morphological features at various spatial scales, efficiently managing the computational load and facilitating a detailed examination of cellular and larger-scale tissue-level details. This method addresses the tissue heterogeneity of HER2 expression by providing a comprehensive view, leading to a blind testing classification accuracy of 84.70%, on a dataset of 523 core images from tissue microarrays. Our automated system, proving reliable as an adjunct pathology tool, has the potential to enhance diagnostic precision and evaluation speed, and might significantly impact cancer treatment planning.
Multiplexed all-optical permutation operations using a reconfigurable diffractive optical network
Feb 04, 2024



Abstract:Large-scale and high-dimensional permutation operations are important for various applications in e.g., telecommunications and encryption. Here, we demonstrate the use of all-optical diffractive computing to execute a set of high-dimensional permutation operations between an input and output field-of-view through layer rotations in a diffractive optical network. In this reconfigurable multiplexed material designed by deep learning, every diffractive layer has four orientations: 0, 90, 180, and 270 degrees. Each unique combination of these rotatable layers represents a distinct rotation state of the diffractive design tailored for a specific permutation operation. Therefore, a K-layer rotatable diffractive material is capable of all-optically performing up to 4^K independent permutation operations. The original input information can be decrypted by applying the specific inverse permutation matrix to output patterns, while applying other inverse operations will lead to loss of information. We demonstrated the feasibility of this reconfigurable multiplexed diffractive design by approximating 256 randomly selected permutation matrices using K=4 rotatable diffractive layers. We also experimentally validated this reconfigurable diffractive network using terahertz radiation and 3D-printed diffractive layers, providing a decent match to our numerical results. The presented rotation-multiplexed diffractive processor design is particularly useful due to its mechanical reconfigurability, offering multifunctional representation through a single fabrication process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge