Tairan Liu
Deep Learning-based Detection of Bacterial Swarm Motion Using a Single Image
Oct 19, 2024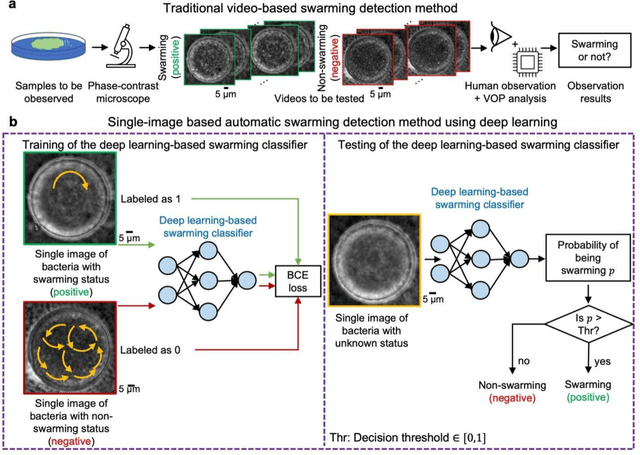
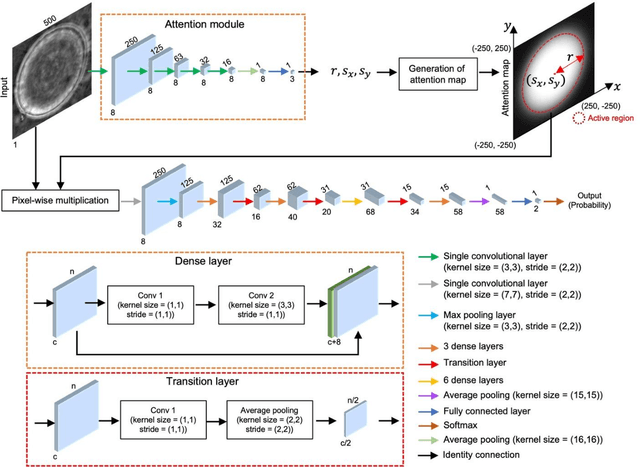
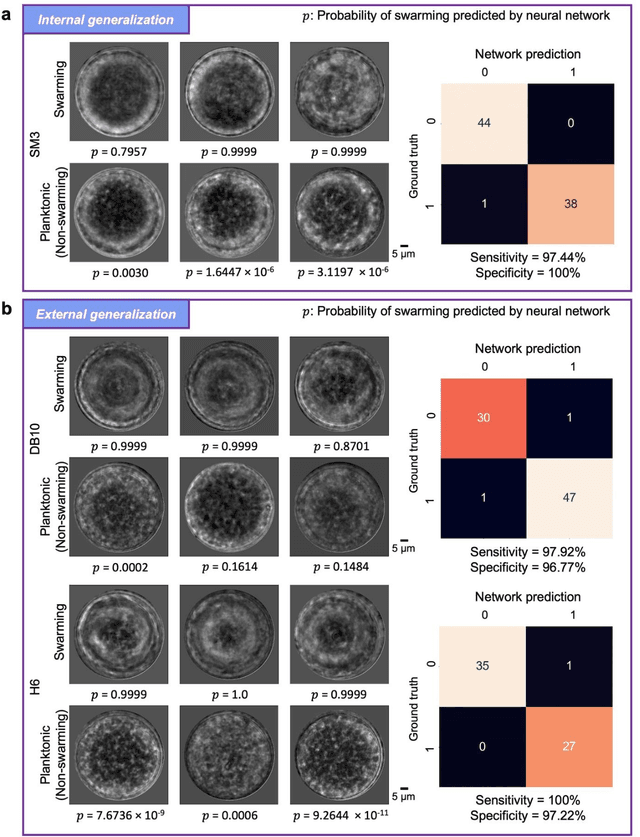
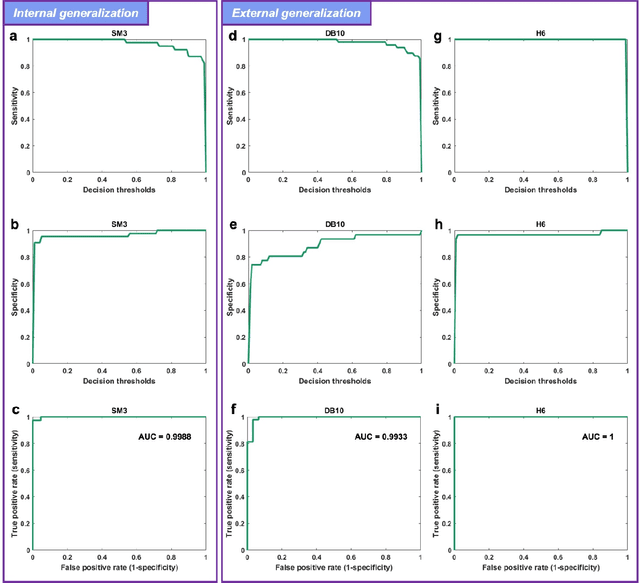
Abstract:Distinguishing between swarming and swimming, the two principal forms of bacterial movement, holds significant conceptual and clinical relevance. This is because bacteria that exhibit swarming capabilities often possess unique properties crucial to the pathogenesis of infectious diseases and may also have therapeutic potential. Here, we report a deep learning-based swarming classifier that rapidly and autonomously predicts swarming probability using a single blurry image. Compared with traditional video-based, manually-processed approaches, our method is particularly suited for high-throughput environments and provides objective, quantitative assessments of swarming probability. The swarming classifier demonstrated in our work was trained on Enterobacter sp. SM3 and showed good performance when blindly tested on new swarming (positive) and swimming (negative) test images of SM3, achieving a sensitivity of 97.44% and a specificity of 100%. Furthermore, this classifier demonstrated robust external generalization capabilities when applied to unseen bacterial species, such as Serratia marcescens DB10 and Citrobacter koseri H6. It blindly achieved a sensitivity of 97.92% and a specificity of 96.77% for DB10, and a sensitivity of 100% and a specificity of 97.22% for H6. This competitive performance indicates the potential to adapt our approach for diagnostic applications through portable devices or even smartphones. This adaptation would facilitate rapid, objective, on-site screening for bacterial swarming motility, potentially enhancing the early detection and treatment assessment of various diseases, including inflammatory bowel diseases (IBD) and urinary tract infections (UTI).
Label-free evaluation of lung and heart transplant biopsies using virtual staining
Sep 09, 2024



Abstract:Organ transplantation serves as the primary therapeutic strategy for end-stage organ failures. However, allograft rejection is a common complication of organ transplantation. Histological assessment is essential for the timely detection and diagnosis of transplant rejection and remains the gold standard. Nevertheless, the traditional histochemical staining process is time-consuming, costly, and labor-intensive. Here, we present a panel of virtual staining neural networks for lung and heart transplant biopsies, which digitally convert autofluorescence microscopic images of label-free tissue sections into their brightfield histologically stained counterparts, bypassing the traditional histochemical staining process. Specifically, we virtually generated Hematoxylin and Eosin (H&E), Masson's Trichrome (MT), and Elastic Verhoeff-Van Gieson (EVG) stains for label-free transplant lung tissue, along with H&E and MT stains for label-free transplant heart tissue. Subsequent blind evaluations conducted by three board-certified pathologists have confirmed that the virtual staining networks consistently produce high-quality histology images with high color uniformity, closely resembling their well-stained histochemical counterparts across various tissue features. The use of virtually stained images for the evaluation of transplant biopsies achieved comparable diagnostic outcomes to those obtained via traditional histochemical staining, with a concordance rate of 82.4% for lung samples and 91.7% for heart samples. Moreover, virtual staining models create multiple stains from the same autofluorescence input, eliminating structural mismatches observed between adjacent sections stained in the traditional workflow, while also saving tissue, expert time, and staining costs.
Virtual histological staining of unlabeled autopsy tissue
Aug 02, 2023Abstract:Histological examination is a crucial step in an autopsy; however, the traditional histochemical staining of post-mortem samples faces multiple challenges, including the inferior staining quality due to autolysis caused by delayed fixation of cadaver tissue, as well as the resource-intensive nature of chemical staining procedures covering large tissue areas, which demand substantial labor, cost, and time. These challenges can become more pronounced during global health crises when the availability of histopathology services is limited, resulting in further delays in tissue fixation and more severe staining artifacts. Here, we report the first demonstration of virtual staining of autopsy tissue and show that a trained neural network can rapidly transform autofluorescence images of label-free autopsy tissue sections into brightfield equivalent images that match hematoxylin and eosin (H&E) stained versions of the same samples, eliminating autolysis-induced severe staining artifacts inherent in traditional histochemical staining of autopsied tissue. Our virtual H&E model was trained using >0.7 TB of image data and a data-efficient collaboration scheme that integrates the virtual staining network with an image registration network. The trained model effectively accentuated nuclear, cytoplasmic and extracellular features in new autopsy tissue samples that experienced severe autolysis, such as COVID-19 samples never seen before, where the traditional histochemical staining failed to provide consistent staining quality. This virtual autopsy staining technique can also be extended to necrotic tissue, and can rapidly and cost-effectively generate artifact-free H&E stains despite severe autolysis and cell death, also reducing labor, cost and infrastructure requirements associated with the standard histochemical staining.
eFIN: Enhanced Fourier Imager Network for generalizable autofocusing and pixel super-resolution in holographic imaging
Jan 09, 2023Abstract:The application of deep learning techniques has greatly enhanced holographic imaging capabilities, leading to improved phase recovery and image reconstruction. Here, we introduce a deep neural network termed enhanced Fourier Imager Network (eFIN) as a highly generalizable framework for hologram reconstruction with pixel super-resolution and image autofocusing. Through holographic microscopy experiments involving lung, prostate and salivary gland tissue sections and Papanicolau (Pap) smears, we demonstrate that eFIN has a superior image reconstruction quality and exhibits external generalization to new types of samples never seen during the training phase. This network achieves a wide autofocusing axial range of 0.35 mm, with the capability to accurately predict the hologram axial distances by physics-informed learning. eFIN enables 3x pixel super-resolution imaging and increases the space-bandwidth product of the reconstructed images by 9-fold with almost no performance loss, which allows for significant time savings in holographic imaging and data processing steps. Our results showcase the advancements of eFIN in pushing the boundaries of holographic imaging for various applications in e.g., quantitative phase imaging and label-free microscopy.
GedankenNet: Self-supervised learning of hologram reconstruction using physics consistency
Sep 17, 2022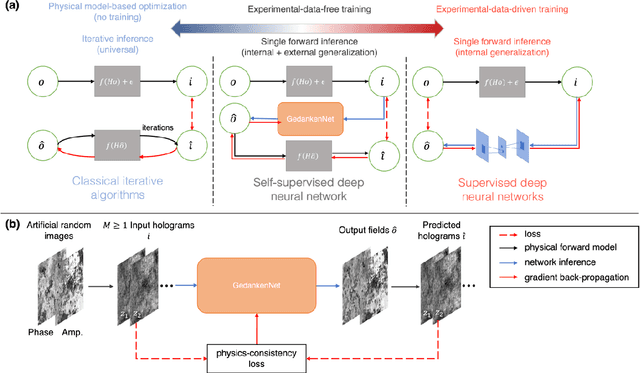
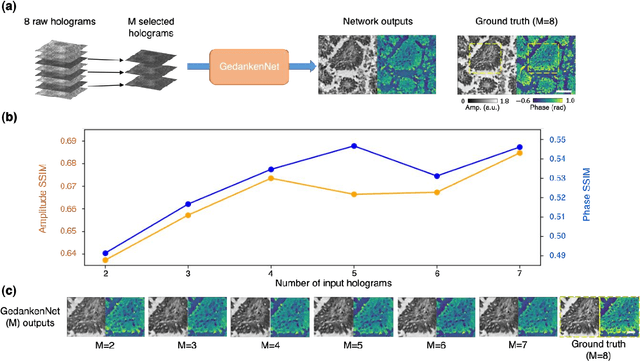

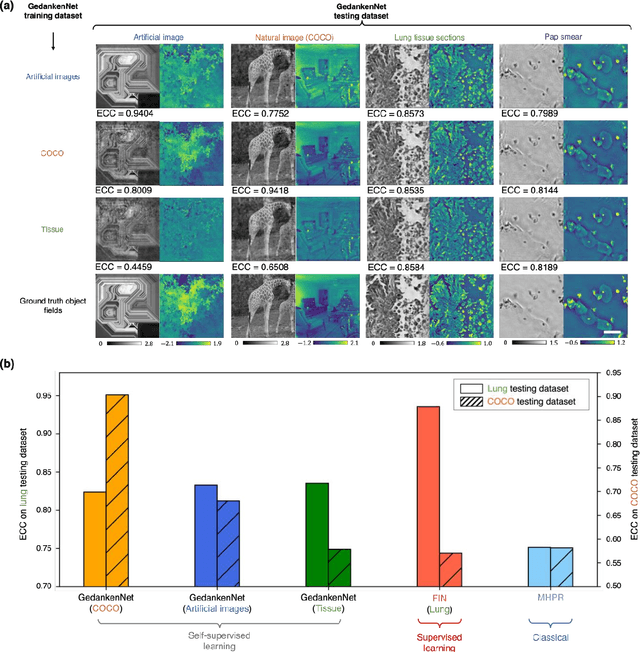
Abstract:The past decade has witnessed transformative applications of deep learning in various computational imaging, sensing and microscopy tasks. Due to the supervised learning schemes employed, most of these methods depend on large-scale, diverse, and labeled training data. The acquisition and preparation of such training image datasets are often laborious and costly, also leading to biased estimation and limited generalization to new types of samples. Here, we report a self-supervised learning model, termed GedankenNet, that eliminates the need for labeled or experimental training data, and demonstrate its effectiveness and superior generalization on hologram reconstruction tasks. Without prior knowledge about the sample types to be imaged, the self-supervised learning model was trained using a physics-consistency loss and artificial random images that are synthetically generated without any experiments or resemblance to real-world samples. After its self-supervised training, GedankenNet successfully generalized to experimental holograms of various unseen biological samples, reconstructing the phase and amplitude images of different types of objects using experimentally acquired test holograms. Without access to experimental data or the knowledge of real samples of interest or their spatial features, GedankenNet's self-supervised learning achieved complex-valued image reconstructions that are consistent with the Maxwell's equations, meaning that its output inference and object solutions accurately represent the wave propagation in free-space. This self-supervised learning of image reconstruction tasks opens up new opportunities for various inverse problems in holography, microscopy and computational imaging fields.
Virtual impactor-based label-free bio-aerosol detection using holography and deep learning
Aug 30, 2022
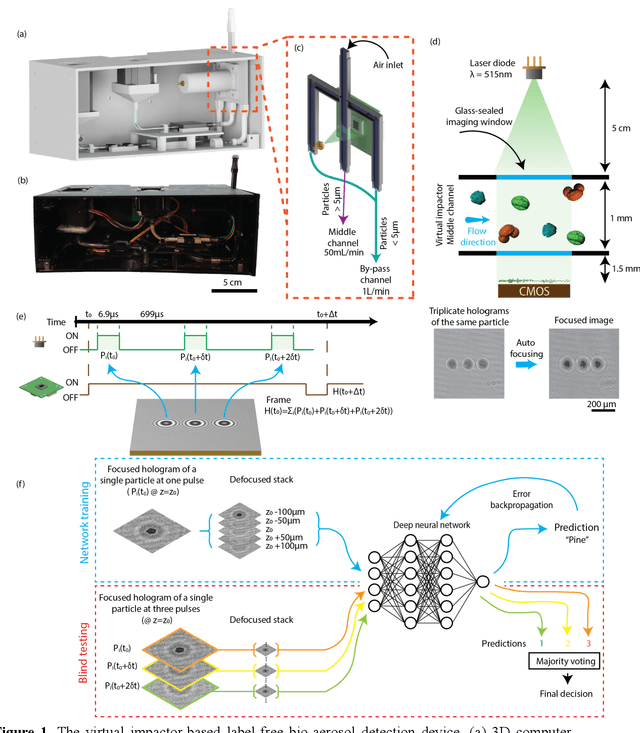
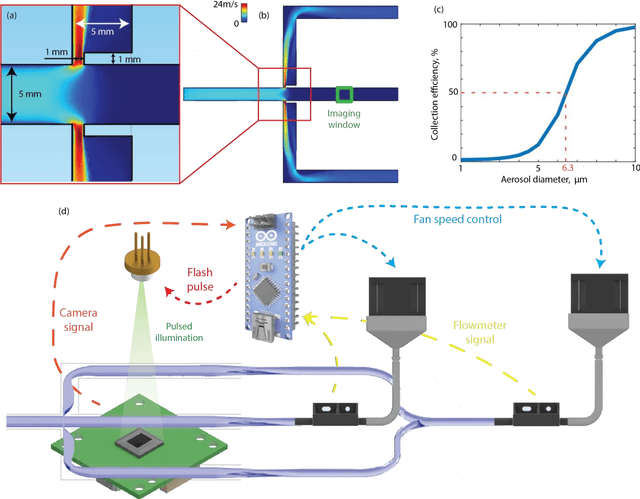
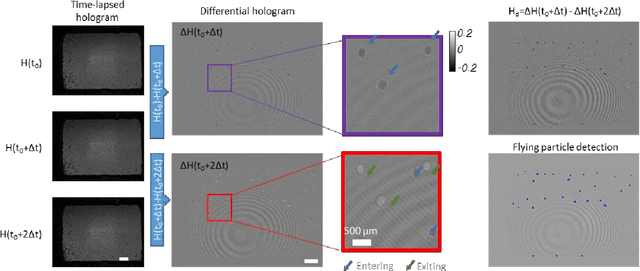
Abstract:Exposure to bio-aerosols such as mold spores and pollen can lead to adverse health effects. There is a need for a portable and cost-effective device for long-term monitoring and quantification of various bio-aerosols. To address this need, we present a mobile and cost-effective label-free bio-aerosol sensor that takes holographic images of flowing particulate matter concentrated by a virtual impactor, which selectively slows down and guides particles larger than ~6 microns to fly through an imaging window. The flowing particles are illuminated by a pulsed laser diode, casting their inline holograms on a CMOS image sensor in a lens-free mobile imaging device. The illumination contains three short pulses with a negligible shift of the flowing particle within one pulse, and triplicate holograms of the same particle are recorded at a single frame before it exits the imaging field-of-view, revealing different perspectives of each particle. The particles within the virtual impactor are localized through a differential detection scheme, and a deep neural network classifies the aerosol type in a label-free manner, based on the acquired holographic images. We demonstrated the success of this mobile bio-aerosol detector with a virtual impactor using different types of pollen (i.e., bermuda, elm, oak, pine, sycamore, and wheat) and achieved a blind classification accuracy of 92.91%. This mobile and cost-effective device weighs ~700 g and can be used for label-free sensing and quantification of various bio-aerosols over extended periods since it is based on a cartridge-free virtual impactor that does not capture or immobilize particulate matter.
Virtual stain transfer in histology via cascaded deep neural networks
Jul 14, 2022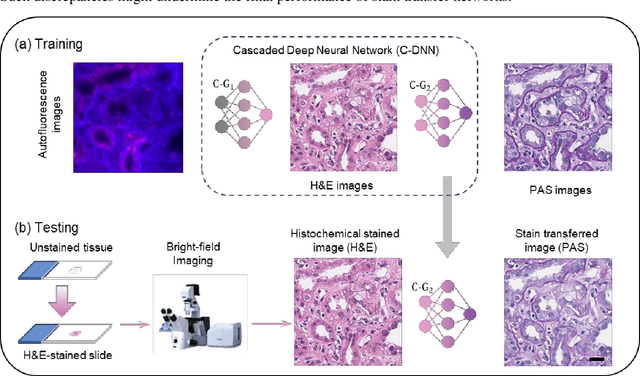
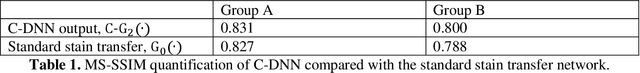
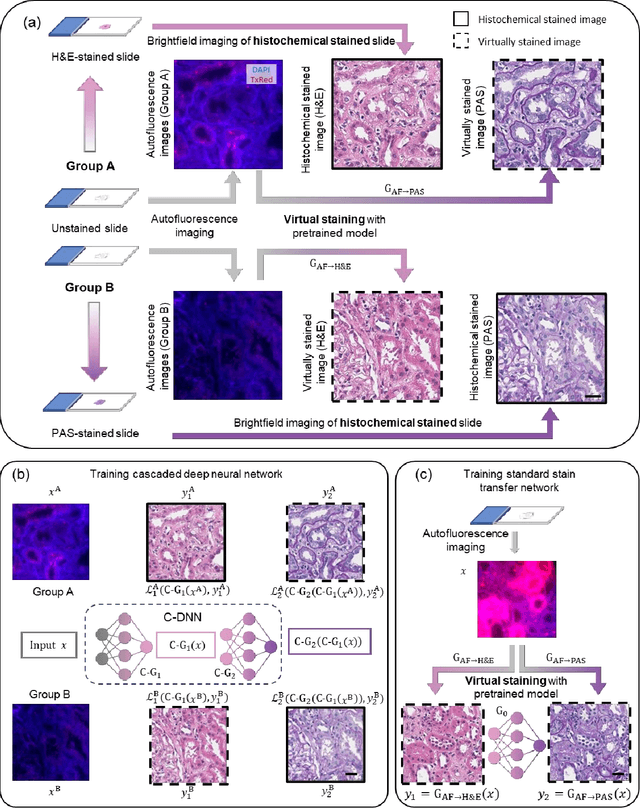
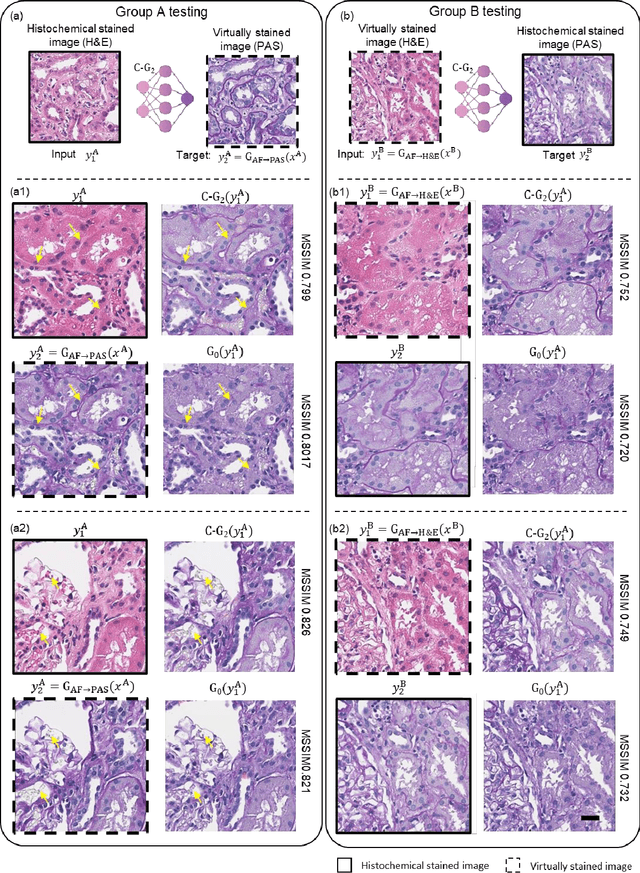
Abstract:Pathological diagnosis relies on the visual inspection of histologically stained thin tissue specimens, where different types of stains are applied to bring contrast to and highlight various desired histological features. However, the destructive histochemical staining procedures are usually irreversible, making it very difficult to obtain multiple stains on the same tissue section. Here, we demonstrate a virtual stain transfer framework via a cascaded deep neural network (C-DNN) to digitally transform hematoxylin and eosin (H&E) stained tissue images into other types of histological stains. Unlike a single neural network structure which only takes one stain type as input to digitally output images of another stain type, C-DNN first uses virtual staining to transform autofluorescence microscopy images into H&E and then performs stain transfer from H&E to the domain of the other stain in a cascaded manner. This cascaded structure in the training phase allows the model to directly exploit histochemically stained image data on both H&E and the target special stain of interest. This advantage alleviates the challenge of paired data acquisition and improves the image quality and color accuracy of the virtual stain transfer from H&E to another stain. We validated the superior performance of this C-DNN approach using kidney needle core biopsy tissue sections and successfully transferred the H&E-stained tissue images into virtual PAS (periodic acid-Schiff) stain. This method provides high-quality virtual images of special stains using existing, histochemically stained slides and creates new opportunities in digital pathology by performing highly accurate stain-to-stain transformations.
Virtual staining of defocused autofluorescence images of unlabeled tissue using deep neural networks
Jul 06, 2022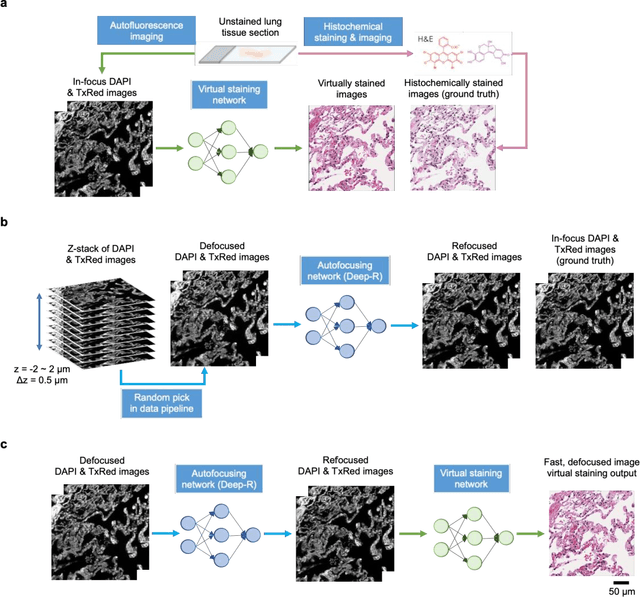
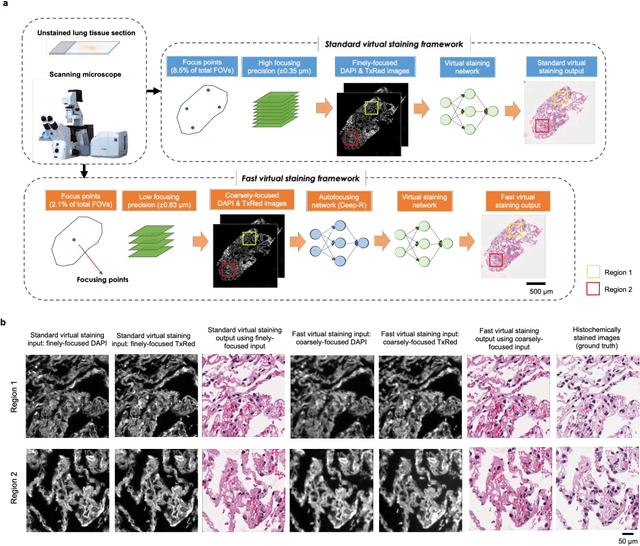
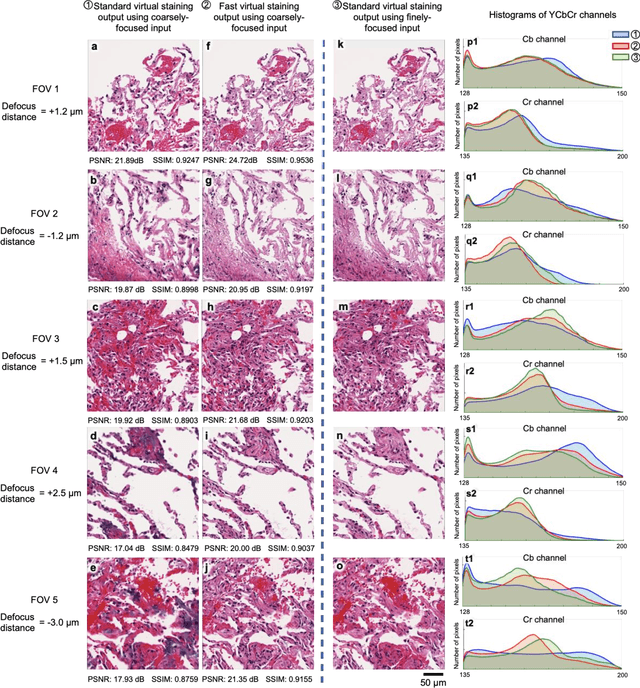
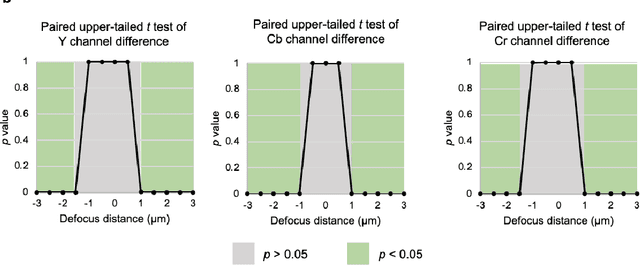
Abstract:Deep learning-based virtual staining was developed to introduce image contrast to label-free tissue sections, digitally matching the histological staining, which is time-consuming, labor-intensive, and destructive to tissue. Standard virtual staining requires high autofocusing precision during the whole slide imaging of label-free tissue, which consumes a significant portion of the total imaging time and can lead to tissue photodamage. Here, we introduce a fast virtual staining framework that can stain defocused autofluorescence images of unlabeled tissue, achieving equivalent performance to virtual staining of in-focus label-free images, also saving significant imaging time by lowering the microscope's autofocusing precision. This framework incorporates a virtual-autofocusing neural network to digitally refocus the defocused images and then transforms the refocused images into virtually stained images using a successive network. These cascaded networks form a collaborative inference scheme: the virtual staining model regularizes the virtual-autofocusing network through a style loss during the training. To demonstrate the efficacy of this framework, we trained and blindly tested these networks using human lung tissue. Using 4x fewer focus points with 2x lower focusing precision, we successfully transformed the coarsely-focused autofluorescence images into high-quality virtually stained H&E images, matching the standard virtual staining framework that used finely-focused autofluorescence input images. Without sacrificing the staining quality, this framework decreases the total image acquisition time needed for virtual staining of a label-free whole-slide image (WSI) by ~32%, together with a ~89% decrease in the autofocusing time, and has the potential to eliminate the laborious and costly histochemical staining process in pathology.
Stain-free, rapid, and quantitative viral plaque assay using deep learning and holography
Jun 30, 2022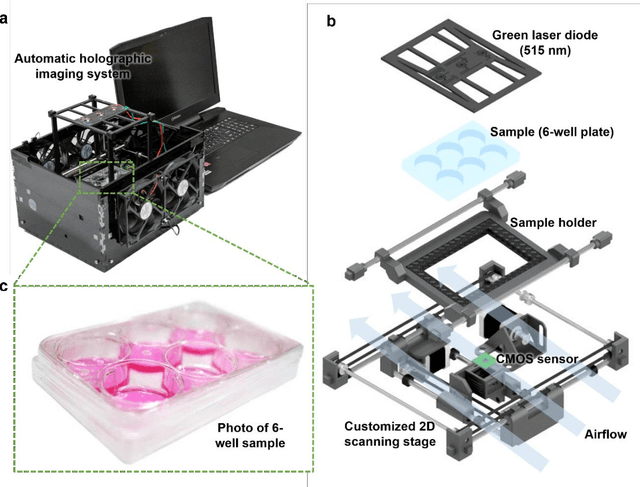
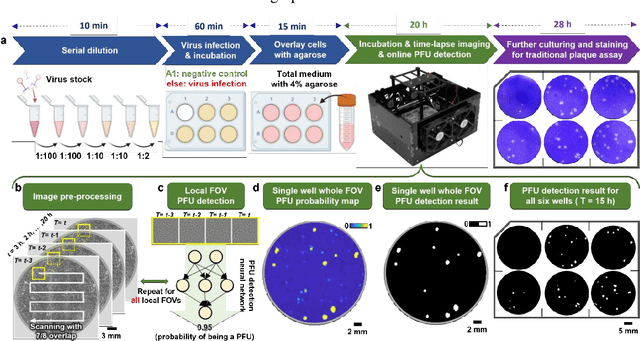
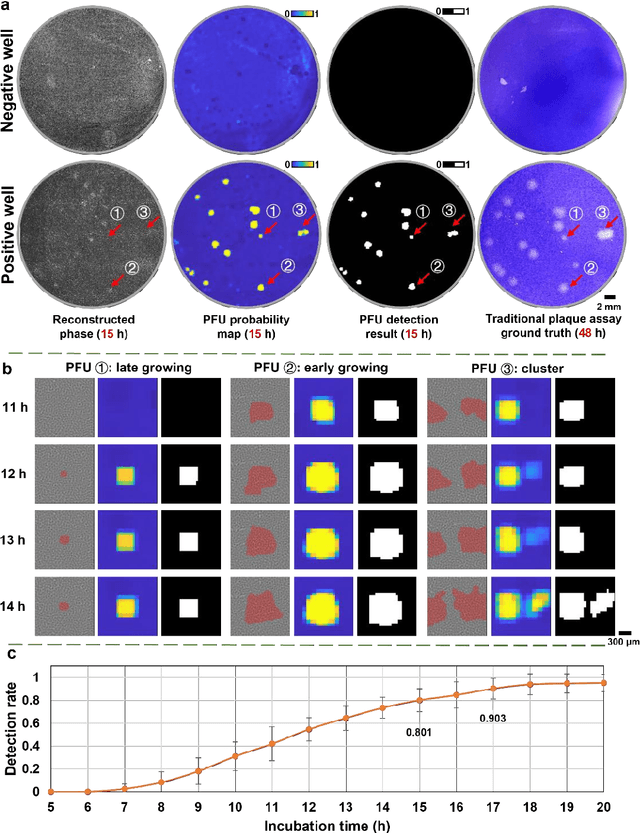
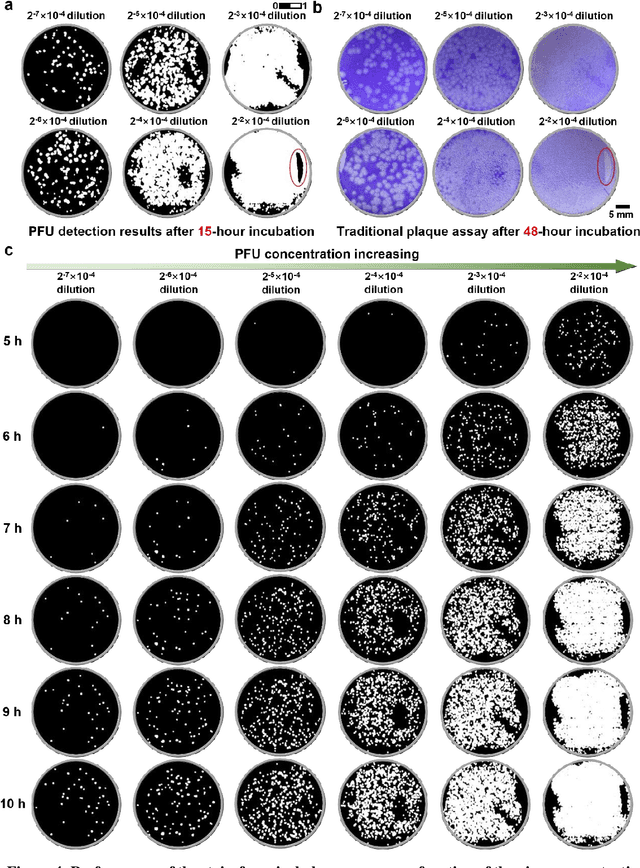
Abstract:Plaque assay is the gold standard method for quantifying the concentration of replication-competent lytic virions. Expediting and automating viral plaque assays will significantly benefit clinical diagnosis, vaccine development, and the production of recombinant proteins or antiviral agents. Here, we present a rapid and stain-free quantitative viral plaque assay using lensfree holographic imaging and deep learning. This cost-effective, compact, and automated device significantly reduces the incubation time needed for traditional plaque assays while preserving their advantages over other virus quantification methods. This device captures ~0.32 Giga-pixel/hour phase information of the objects per test well, covering an area of ~30x30 mm^2, in a label-free manner, eliminating staining entirely. We demonstrated the success of this computational method using Vero E6 cells and vesicular stomatitis virus. Using a neural network, this stain-free device automatically detected the first cell lysing events due to the viral replication as early as 5 hours after the incubation, and achieved >90% detection rate for the plaque-forming units (PFUs) with 100% specificity in <20 hours, providing major time savings compared to the traditional plaque assays that take ~48 hours or more. This data-driven plaque assay also offers the capability of quantifying the infected area of the cell monolayer, performing automated counting and quantification of PFUs and virus-infected areas over a 10-fold larger dynamic range of virus concentration than standard viral plaque assays. This compact, low-cost, automated PFU quantification device can be broadly used in virology research, vaccine development, and clinical applications
Deep Learning-enabled Detection and Classification of Bacterial Colonies using a Thin Film Transistor (TFT) Image Sensor
May 07, 2022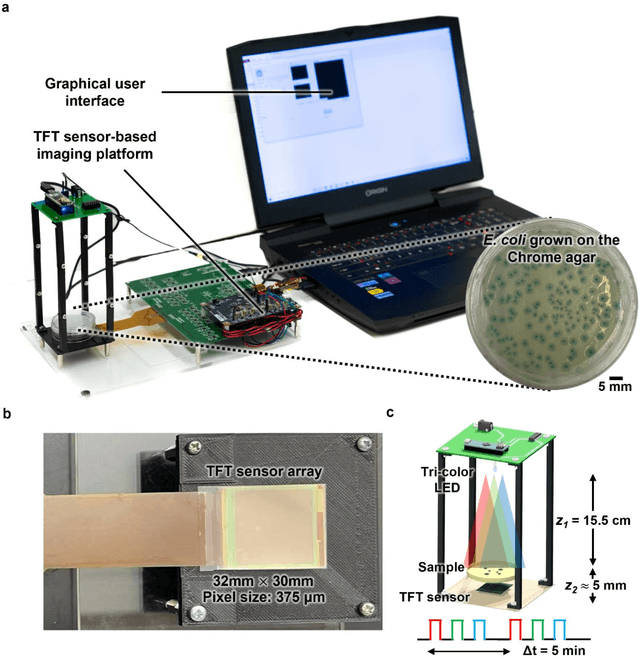
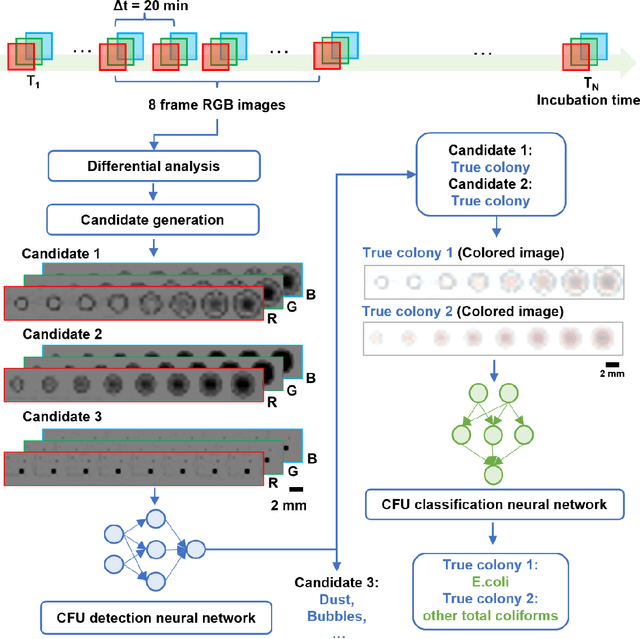
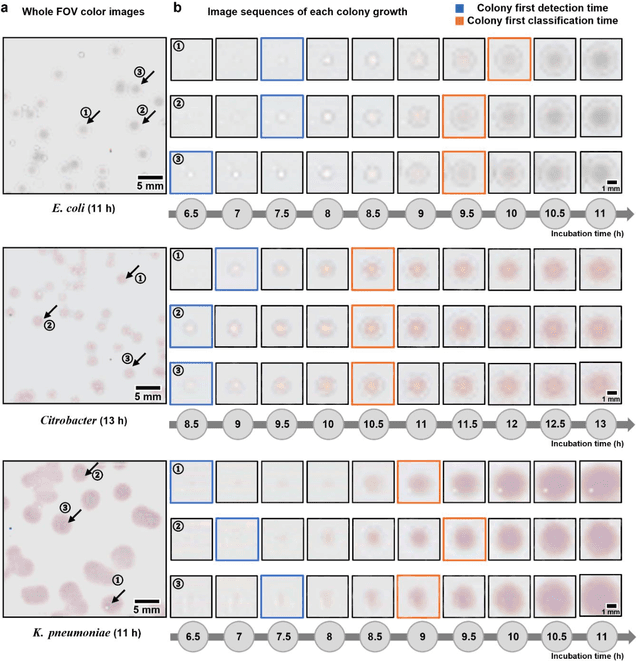
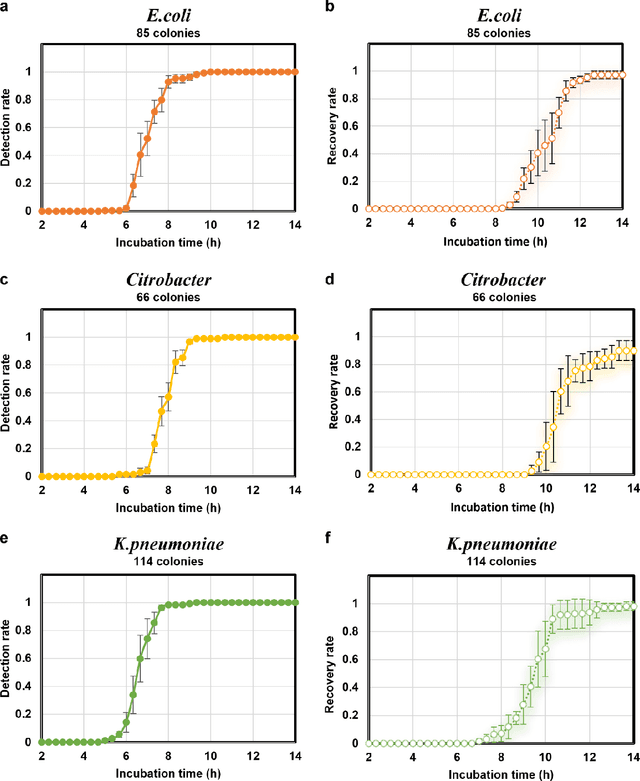
Abstract:Early detection and identification of pathogenic bacteria such as Escherichia coli (E. coli) is an essential task for public health. The conventional culture-based methods for bacterial colony detection usually take >24 hours to get the final read-out. Here, we demonstrate a bacterial colony-forming-unit (CFU) detection system exploiting a thin-film-transistor (TFT)-based image sensor array that saves ~12 hours compared to the Environmental Protection Agency (EPA)-approved methods. To demonstrate the efficacy of this CFU detection system, a lensfree imaging modality was built using the TFT image sensor with a sample field-of-view of ~10 cm^2. Time-lapse images of bacterial colonies cultured on chromogenic agar plates were automatically collected at 5-minute intervals. Two deep neural networks were used to detect and count the growing colonies and identify their species. When blindly tested with 265 colonies of E. coli and other coliform bacteria (i.e., Citrobacter and Klebsiella pneumoniae), our system reached an average CFU detection rate of 97.3% at 9 hours of incubation and an average recovery rate of 91.6% at ~12 hours. This TFT-based sensor can be applied to various microbiological detection methods. Due to the large scalability, ultra-large field-of-view, and low cost of the TFT-based image sensors, this platform can be integrated with each agar plate to be tested and disposed of after the automated CFU count. The imaging field-of-view of this platform can be cost-effectively increased to >100 cm^2 to provide a massive throughput for CFU detection using, e.g., roll-to-roll manufacturing of TFTs as used in the flexible display industry.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge