Hongda Wang
Stain-free, rapid, and quantitative viral plaque assay using deep learning and holography
Jun 30, 2022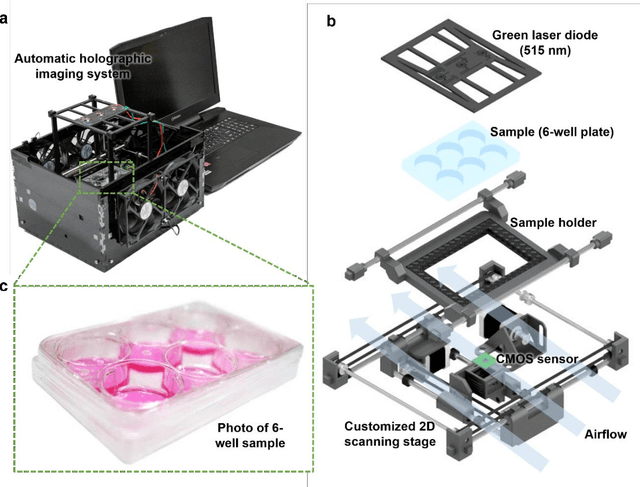
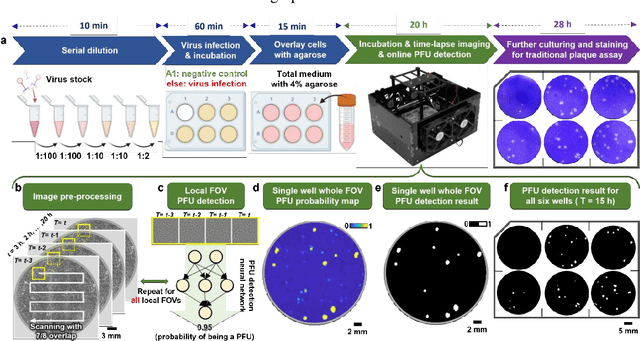
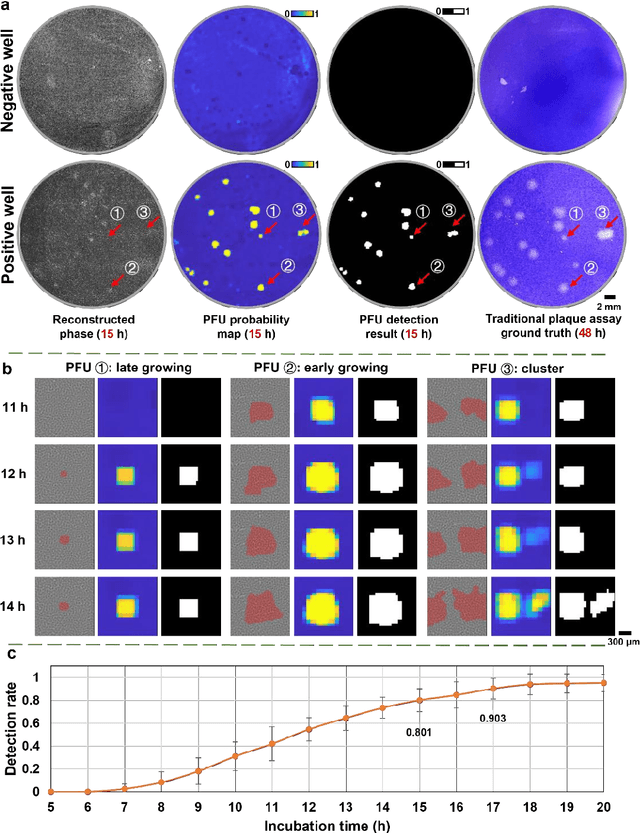
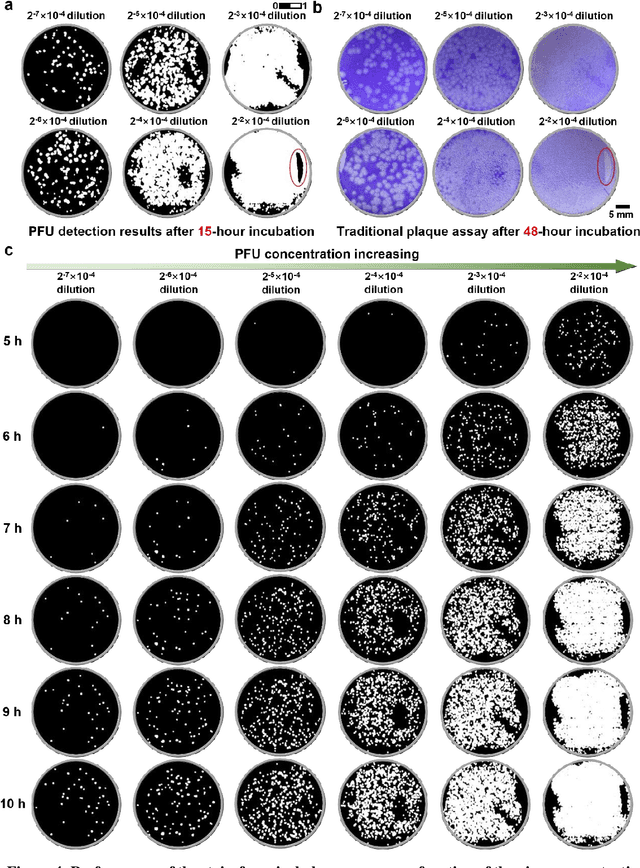
Abstract:Plaque assay is the gold standard method for quantifying the concentration of replication-competent lytic virions. Expediting and automating viral plaque assays will significantly benefit clinical diagnosis, vaccine development, and the production of recombinant proteins or antiviral agents. Here, we present a rapid and stain-free quantitative viral plaque assay using lensfree holographic imaging and deep learning. This cost-effective, compact, and automated device significantly reduces the incubation time needed for traditional plaque assays while preserving their advantages over other virus quantification methods. This device captures ~0.32 Giga-pixel/hour phase information of the objects per test well, covering an area of ~30x30 mm^2, in a label-free manner, eliminating staining entirely. We demonstrated the success of this computational method using Vero E6 cells and vesicular stomatitis virus. Using a neural network, this stain-free device automatically detected the first cell lysing events due to the viral replication as early as 5 hours after the incubation, and achieved >90% detection rate for the plaque-forming units (PFUs) with 100% specificity in <20 hours, providing major time savings compared to the traditional plaque assays that take ~48 hours or more. This data-driven plaque assay also offers the capability of quantifying the infected area of the cell monolayer, performing automated counting and quantification of PFUs and virus-infected areas over a 10-fold larger dynamic range of virus concentration than standard viral plaque assays. This compact, low-cost, automated PFU quantification device can be broadly used in virology research, vaccine development, and clinical applications
Deep Learning-enabled Detection and Classification of Bacterial Colonies using a Thin Film Transistor (TFT) Image Sensor
May 07, 2022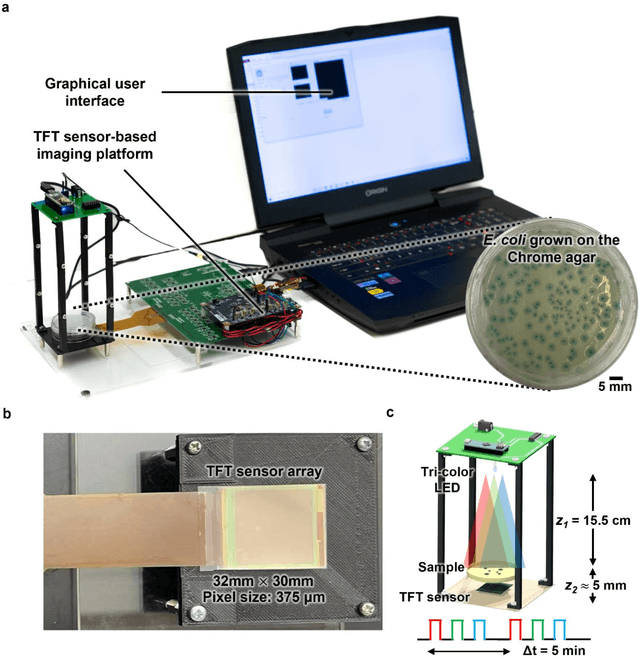
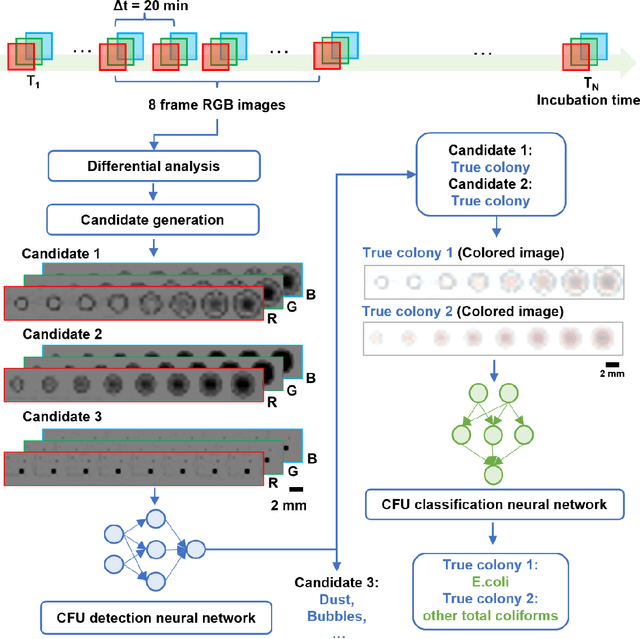
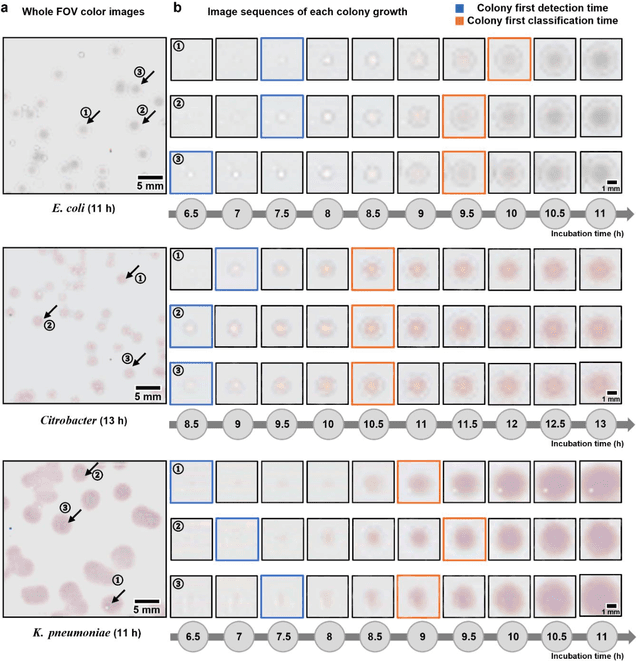
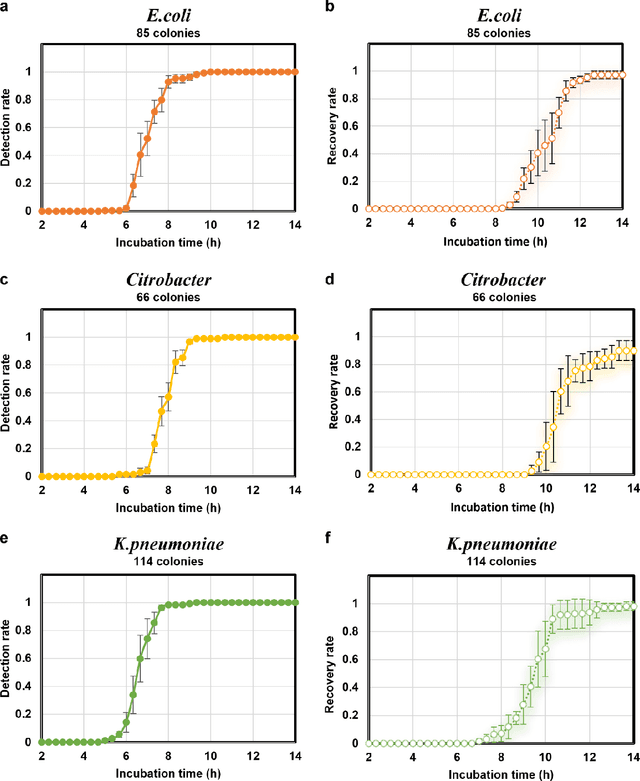
Abstract:Early detection and identification of pathogenic bacteria such as Escherichia coli (E. coli) is an essential task for public health. The conventional culture-based methods for bacterial colony detection usually take >24 hours to get the final read-out. Here, we demonstrate a bacterial colony-forming-unit (CFU) detection system exploiting a thin-film-transistor (TFT)-based image sensor array that saves ~12 hours compared to the Environmental Protection Agency (EPA)-approved methods. To demonstrate the efficacy of this CFU detection system, a lensfree imaging modality was built using the TFT image sensor with a sample field-of-view of ~10 cm^2. Time-lapse images of bacterial colonies cultured on chromogenic agar plates were automatically collected at 5-minute intervals. Two deep neural networks were used to detect and count the growing colonies and identify their species. When blindly tested with 265 colonies of E. coli and other coliform bacteria (i.e., Citrobacter and Klebsiella pneumoniae), our system reached an average CFU detection rate of 97.3% at 9 hours of incubation and an average recovery rate of 91.6% at ~12 hours. This TFT-based sensor can be applied to various microbiological detection methods. Due to the large scalability, ultra-large field-of-view, and low cost of the TFT-based image sensors, this platform can be integrated with each agar plate to be tested and disposed of after the automated CFU count. The imaging field-of-view of this platform can be cost-effectively increased to >100 cm^2 to provide a massive throughput for CFU detection using, e.g., roll-to-roll manufacturing of TFTs as used in the flexible display industry.
Label-free virtual HER2 immunohistochemical staining of breast tissue using deep learning
Dec 08, 2021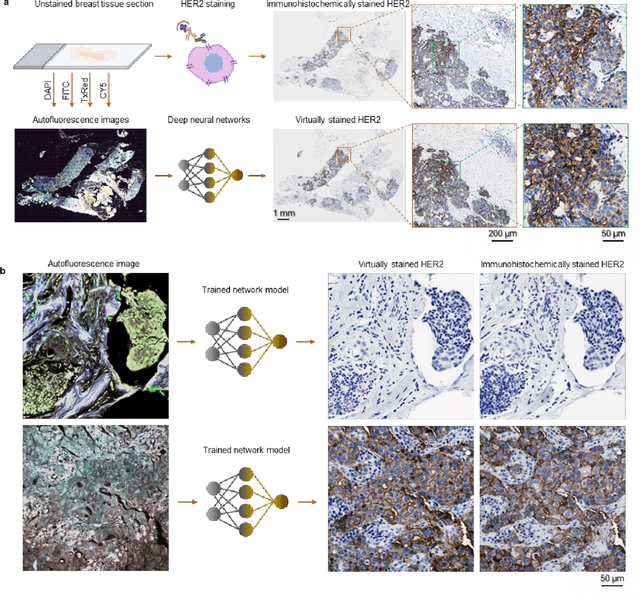
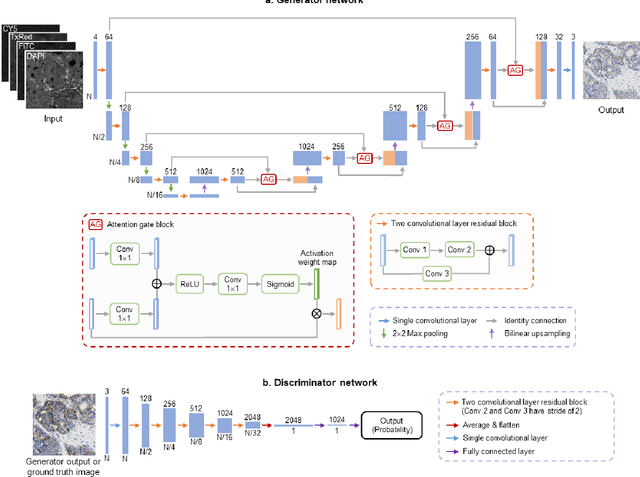
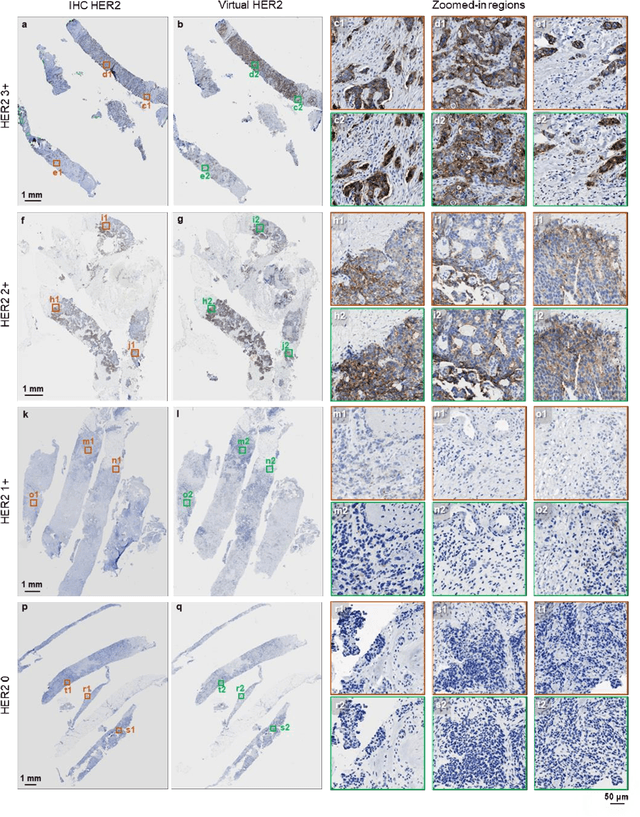
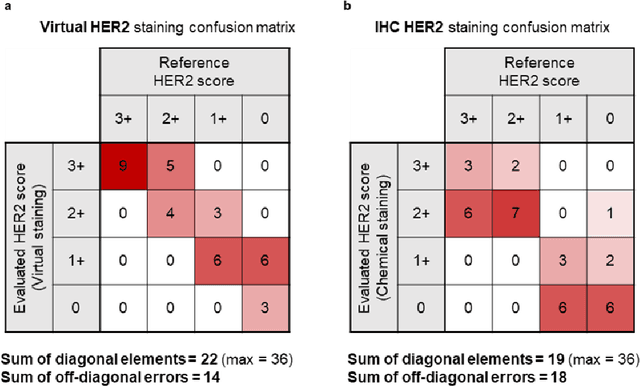
Abstract:The immunohistochemical (IHC) staining of the human epidermal growth factor receptor 2 (HER2) biomarker is widely practiced in breast tissue analysis, preclinical studies and diagnostic decisions, guiding cancer treatment and investigation of pathogenesis. HER2 staining demands laborious tissue treatment and chemical processing performed by a histotechnologist, which typically takes one day to prepare in a laboratory, increasing analysis time and associated costs. Here, we describe a deep learning-based virtual HER2 IHC staining method using a conditional generative adversarial network that is trained to rapidly transform autofluorescence microscopic images of unlabeled/label-free breast tissue sections into bright-field equivalent microscopic images, matching the standard HER2 IHC staining that is chemically performed on the same tissue sections. The efficacy of this virtual HER2 staining framework was demonstrated by quantitative analysis, in which three board-certified breast pathologists blindly graded the HER2 scores of virtually stained and immunohistochemically stained HER2 whole slide images (WSIs) to reveal that the HER2 scores determined by inspecting virtual IHC images are as accurate as their immunohistochemically stained counterparts. A second quantitative blinded study performed by the same diagnosticians further revealed that the virtually stained HER2 images exhibit a comparable staining quality in the level of nuclear detail, membrane clearness, and absence of staining artifacts with respect to their immunohistochemically stained counterparts. This virtual HER2 staining framework bypasses the costly, laborious, and time-consuming IHC staining procedures in laboratory, and can be extended to other types of biomarkers to accelerate the IHC tissue staining used in life sciences and biomedical workflow.
Deep learning-based virtual refocusing of images using an engineered point-spread function
Dec 22, 2020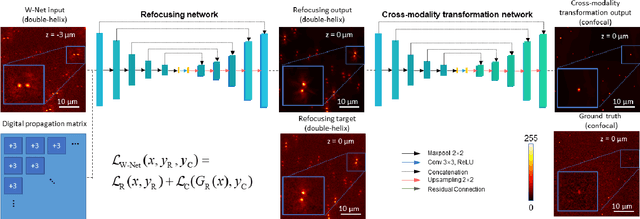
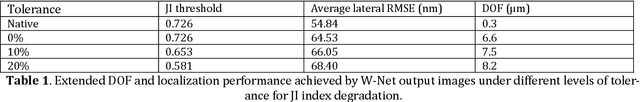
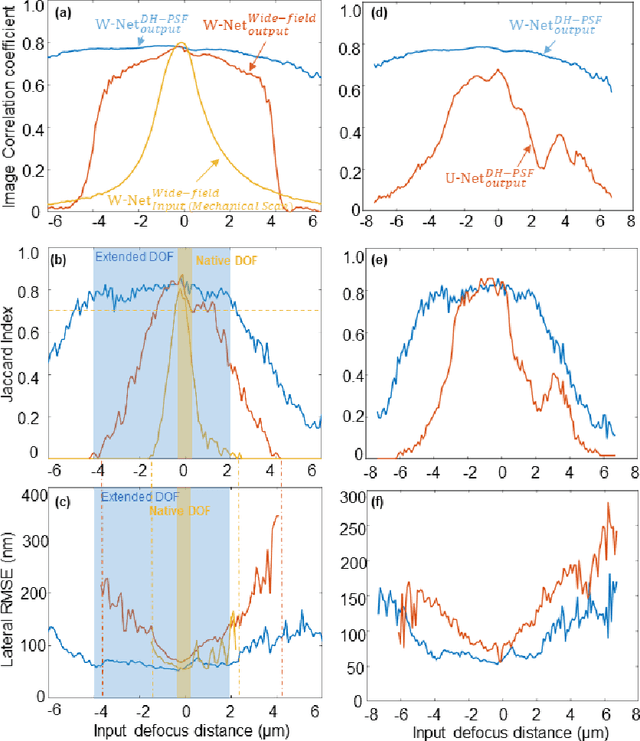
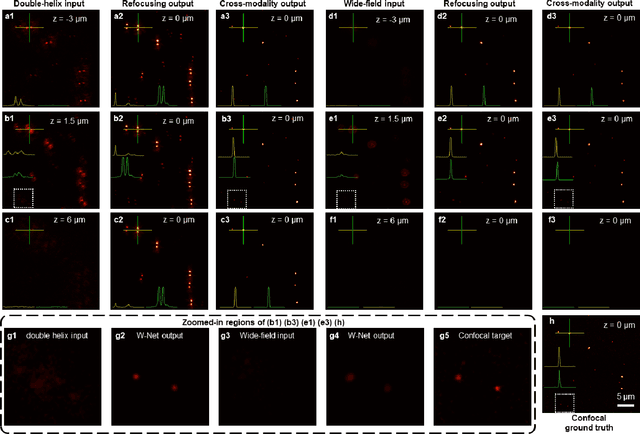
Abstract:We present a virtual image refocusing method over an extended depth of field (DOF) enabled by cascaded neural networks and a double-helix point-spread function (DH-PSF). This network model, referred to as W-Net, is composed of two cascaded generator and discriminator network pairs. The first generator network learns to virtually refocus an input image onto a user-defined plane, while the second generator learns to perform a cross-modality image transformation, improving the lateral resolution of the output image. Using this W-Net model with DH-PSF engineering, we extend the DOF of a fluorescence microscope by ~20-fold. This approach can be applied to develop deep learning-enabled image reconstruction methods for localization microscopy techniques that utilize engineered PSFs to improve their imaging performance, including spatial resolution and volumetric imaging throughput.
Deep learning-based holographic polarization microscopy
Jul 01, 2020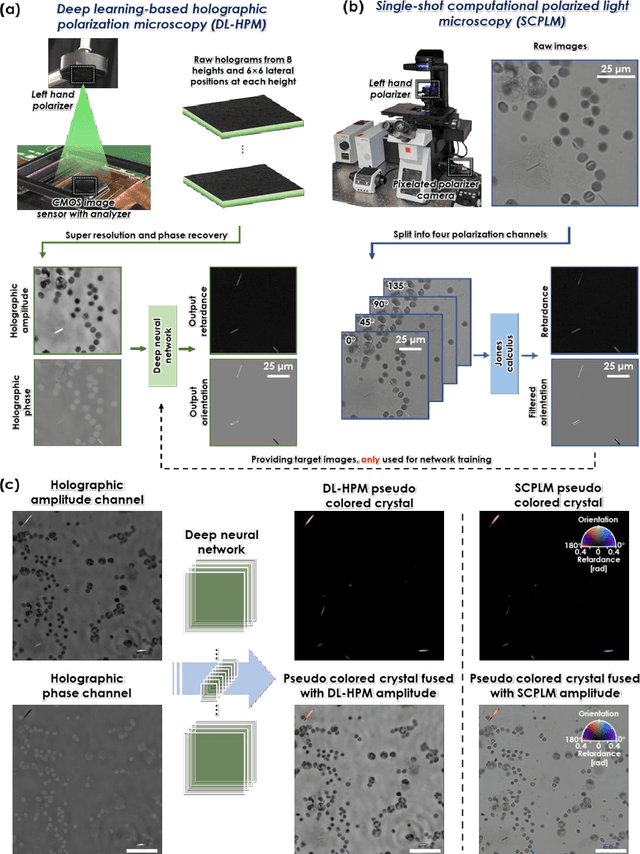
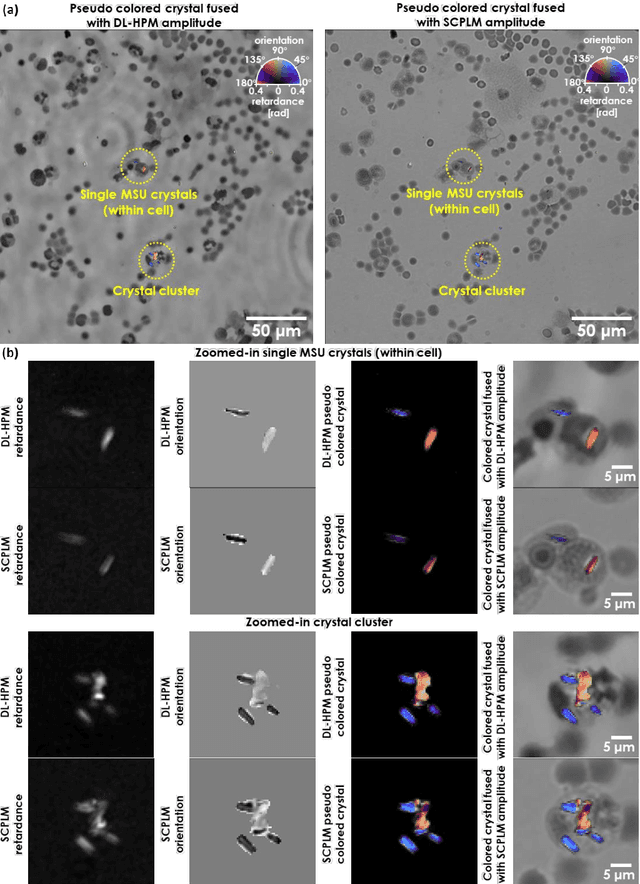
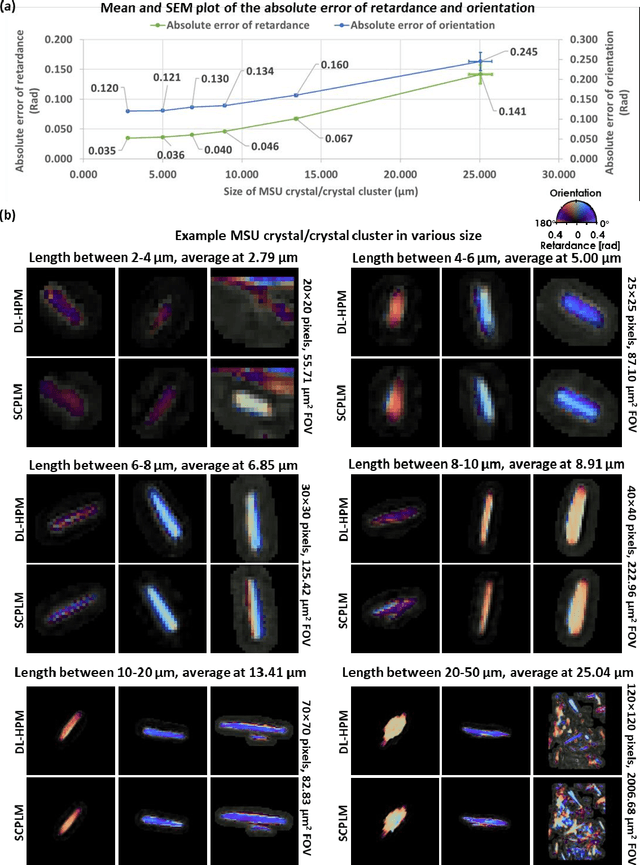
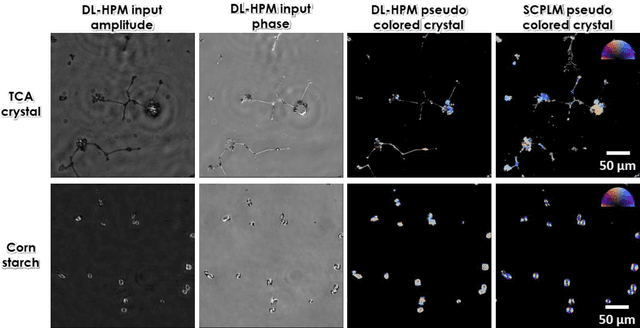
Abstract:Polarized light microscopy provides high contrast to birefringent specimen and is widely used as a diagnostic tool in pathology. However, polarization microscopy systems typically operate by analyzing images collected from two or more light paths in different states of polarization, which lead to relatively complex optical designs, high system costs or experienced technicians being required. Here, we present a deep learning-based holographic polarization microscope that is capable of obtaining quantitative birefringence retardance and orientation information of specimen from a phase recovered hologram, while only requiring the addition of one polarizer/analyzer pair to an existing holographic imaging system. Using a deep neural network, the reconstructed holographic images from a single state of polarization can be transformed into images equivalent to those captured using a single-shot computational polarized light microscope (SCPLM). Our analysis shows that a trained deep neural network can extract the birefringence information using both the sample specific morphological features as well as the holographic amplitude and phase distribution. To demonstrate the efficacy of this method, we tested it by imaging various birefringent samples including e.g., monosodium urate (MSU) and triamcinolone acetonide (TCA) crystals. Our method achieves similar results to SCPLM both qualitatively and quantitatively, and due to its simpler optical design and significantly larger field-of-view, this method has the potential to expand the access to polarization microscopy and its use for medical diagnosis in resource limited settings.
Early-detection and classification of live bacteria using time-lapse coherent imaging and deep learning
Jan 29, 2020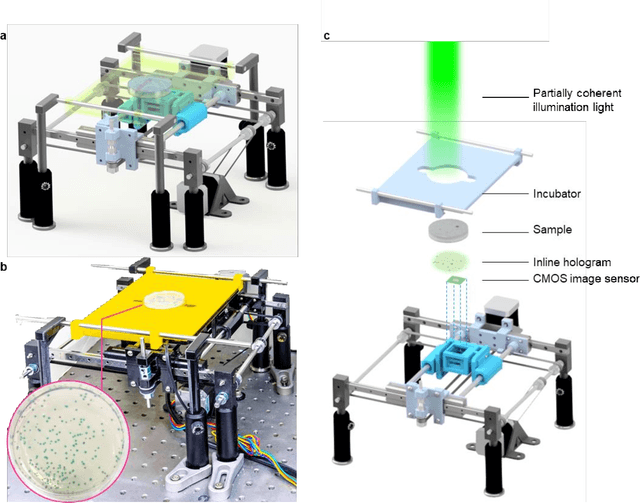
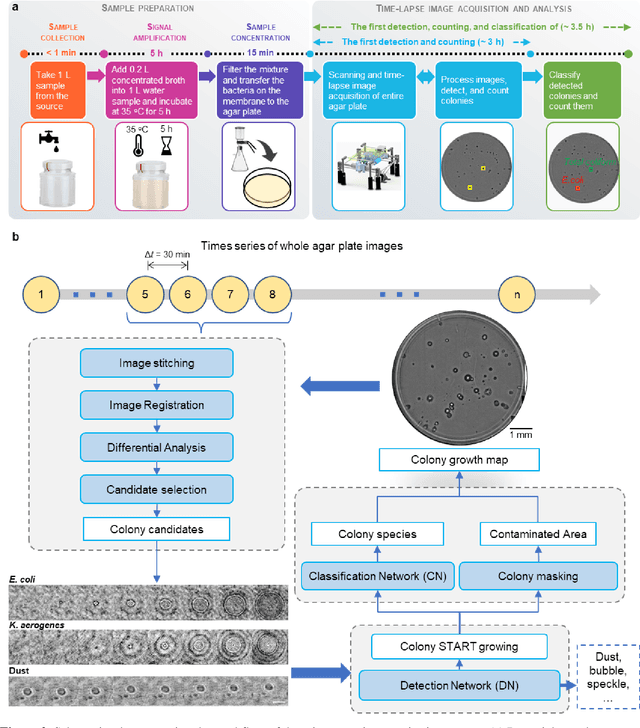
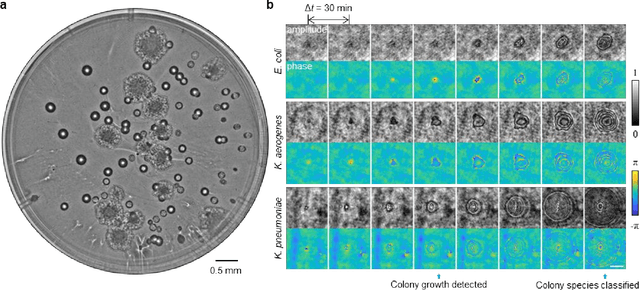
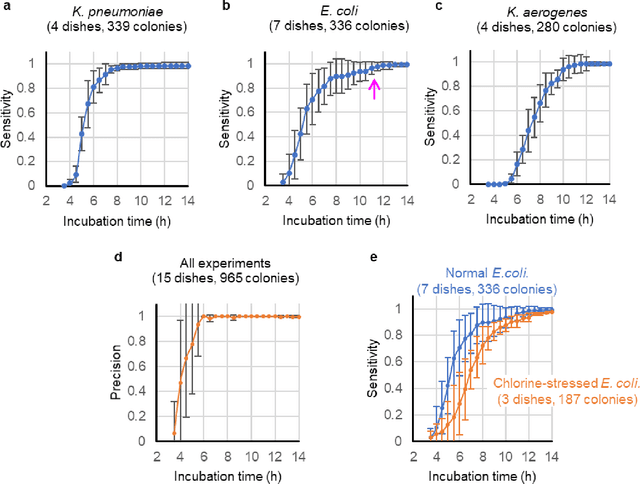
Abstract:We present a computational live bacteria detection system that periodically captures coherent microscopy images of bacterial growth inside a 60 mm diameter agar-plate and analyzes these time-lapsed holograms using deep neural networks for rapid detection of bacterial growth and classification of the corresponding species. The performance of our system was demonstrated by rapid detection of Escherichia coli and total coliform bacteria (i.e., Klebsiella aerogenes and Klebsiella pneumoniae subsp. pneumoniae) in water samples. These results were confirmed against gold-standard culture-based results, shortening the detection time of bacterial growth by >12 h as compared to the Environmental Protection Agency (EPA)-approved analytical methods. Our experiments further confirmed that this method successfully detects 90% of bacterial colonies within 7-10 h (and >95% within 12 h) with a precision of 99.2-100%, and correctly identifies their species in 7.6-12 h with 80% accuracy. Using pre-incubation of samples in growth media, our system achieved a limit of detection (LOD) of ~1 colony forming unit (CFU)/L within 9 h of total test time. This computational bacteria detection and classification platform is highly cost-effective (~$0.6 per test) and high-throughput with a scanning speed of 24 cm2/min over the entire plate surface, making it highly suitable for integration with the existing analytical methods currently used for bacteria detection on agar plates. Powered by deep learning, this automated and cost-effective live bacteria detection platform can be transformative for a wide range of applications in microbiology by significantly reducing the detection time, also automating the identification of colonies, without labeling or the need for an expert.
Three-dimensional propagation and time-reversal of fluorescence images
Jan 31, 2019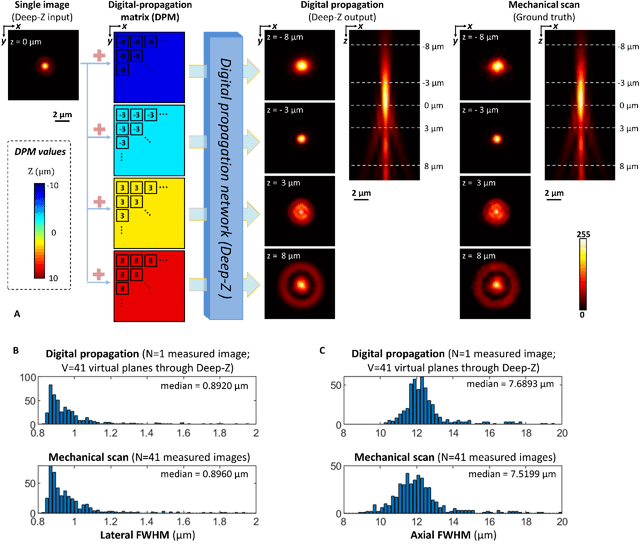
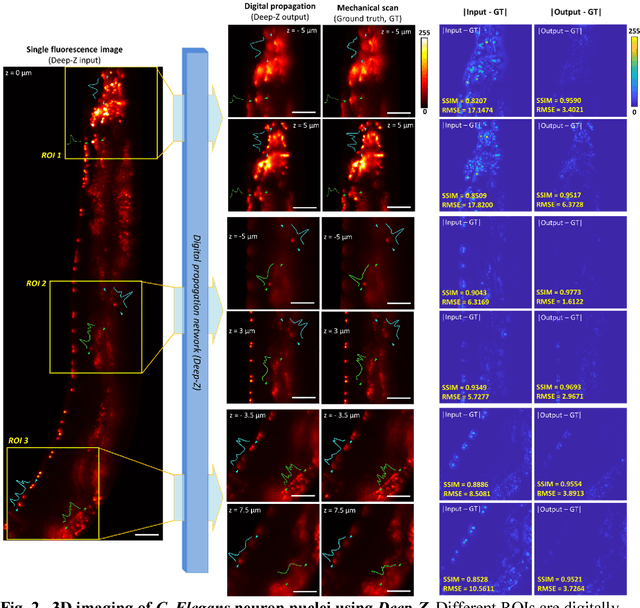
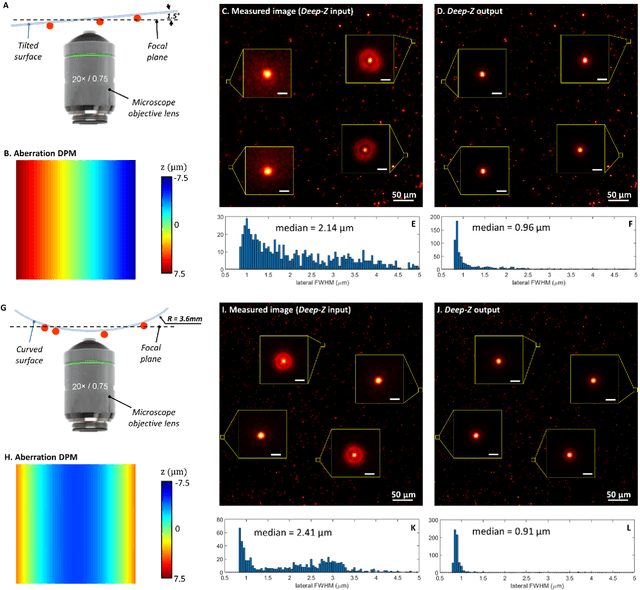
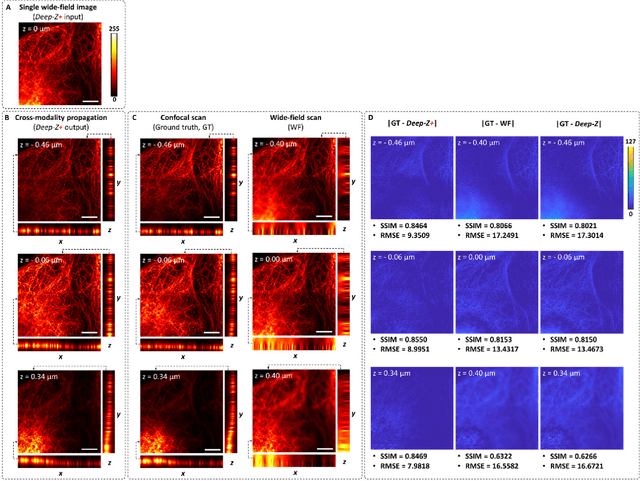
Abstract:Unlike holography, fluorescence microscopy lacks an image propagation and time-reversal framework, which necessitates scanning of fluorescent objects to obtain 3D images. We demonstrate that a neural network can inherently learn the physical laws governing fluorescence wave propagation and time-reversal to enable 3D imaging of fluorescent samples using a single 2D image, without mechanical scanning, additional hardware, or a trade-off of resolution or speed. Using this data-driven framework, we increased the depth-of-field of a microscope by 20-fold, imaged Caenorhabditis elegans neurons in 3D using a single fluorescence image, and digitally propagated fluorescence images onto user-defined 3D surfaces, also correcting various aberrations. Furthermore, this learning-based approach cross-connects different imaging modalities, permitting 3D propagation of a wide-field fluorescence image to match confocal microscopy images acquired at different sample planes.
Deep learning-based virtual histology staining using auto-fluorescence of label-free tissue
Mar 30, 2018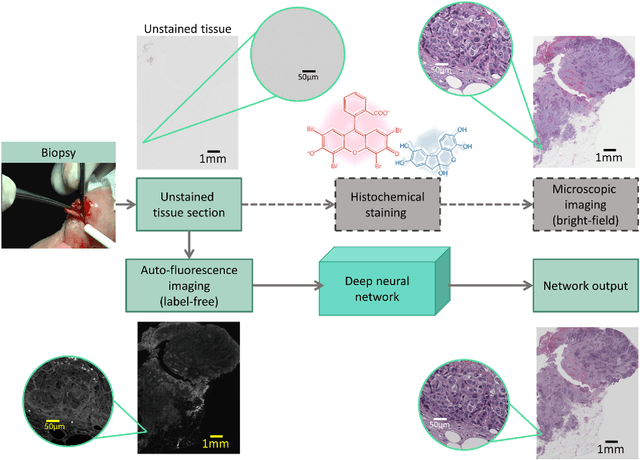
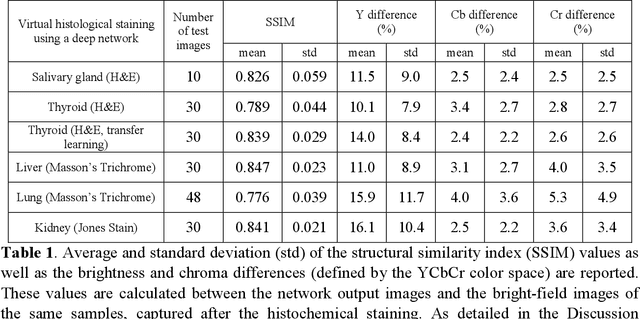
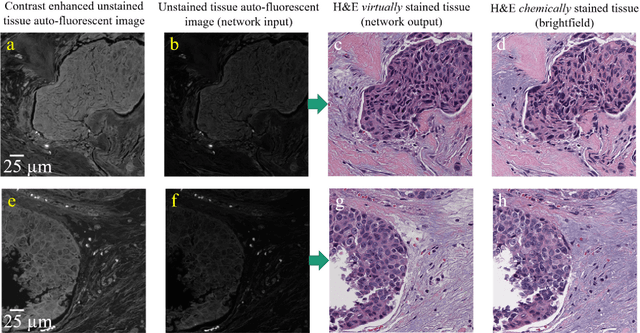
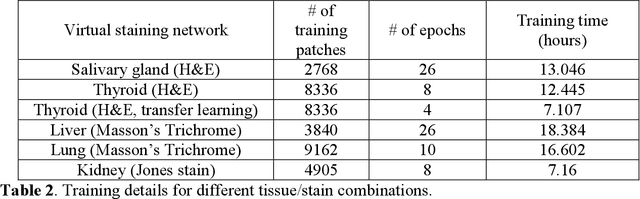
Abstract:Histological analysis of tissue samples is one of the most widely used methods for disease diagnosis. After taking a sample from a patient, it goes through a lengthy and laborious preparation, which stains the tissue to visualize different histological features under a microscope. Here, we demonstrate a label-free approach to create a virtually-stained microscopic image using a single wide-field auto-fluorescence image of an unlabeled tissue sample, bypassing the standard histochemical staining process, saving time and cost. This method is based on deep learning, and uses a convolutional neural network trained using a generative adversarial network model to transform an auto-fluorescence image of an unlabeled tissue section into an image that is equivalent to the bright-field image of the stained-version of the same sample. We validated this method by successfully creating virtually-stained microscopic images of human tissue samples, including sections of salivary gland, thyroid, kidney, liver and lung tissue, also covering three different stains. This label-free virtual-staining method eliminates cumbersome and costly histochemical staining procedures, and would significantly simplify tissue preparation in pathology and histology fields.
Deep learning enhanced mobile-phone microscopy
Dec 12, 2017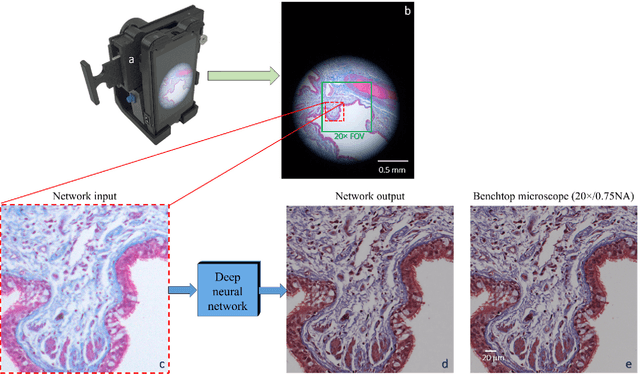
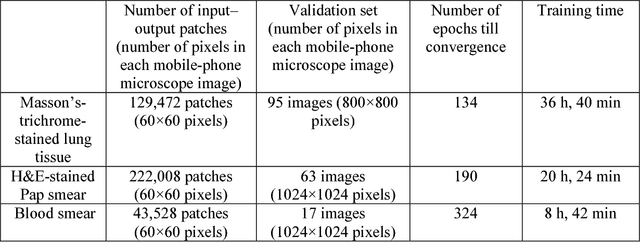
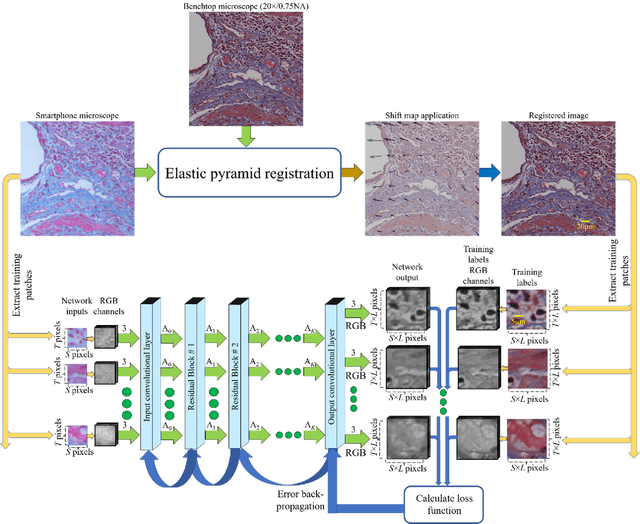
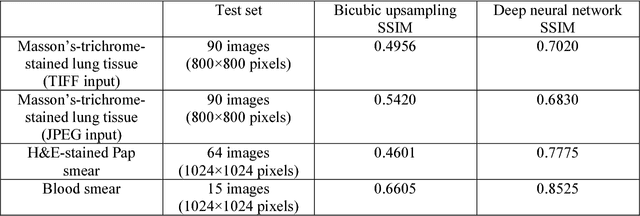
Abstract:Mobile-phones have facilitated the creation of field-portable, cost-effective imaging and sensing technologies that approach laboratory-grade instrument performance. However, the optical imaging interfaces of mobile-phones are not designed for microscopy and produce spatial and spectral distortions in imaging microscopic specimens. Here, we report on the use of deep learning to correct such distortions introduced by mobile-phone-based microscopes, facilitating the production of high-resolution, denoised and colour-corrected images, matching the performance of benchtop microscopes with high-end objective lenses, also extending their limited depth-of-field. After training a convolutional neural network, we successfully imaged various samples, including blood smears, histopathology tissue sections, and parasites, where the recorded images were highly compressed to ease storage and transmission for telemedicine applications. This method is applicable to other low-cost, aberrated imaging systems, and could offer alternatives for costly and bulky microscopes, while also providing a framework for standardization of optical images for clinical and biomedical applications.
Deep Learning Microscopy
May 12, 2017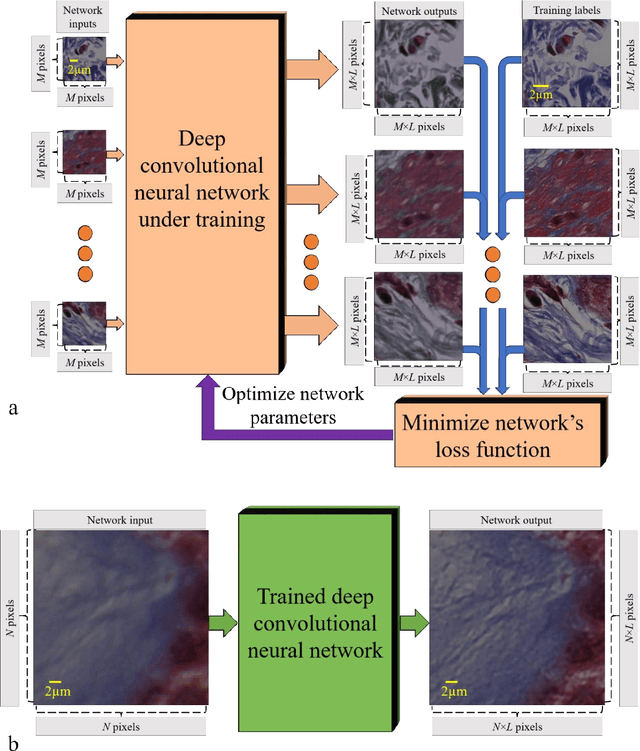
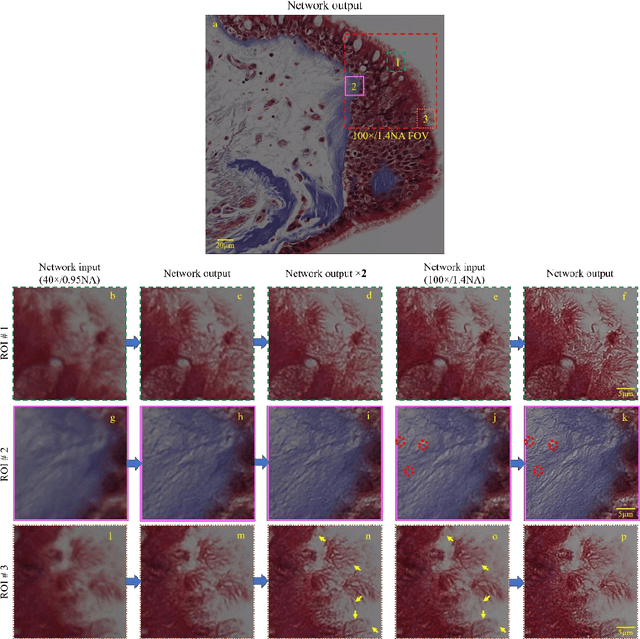
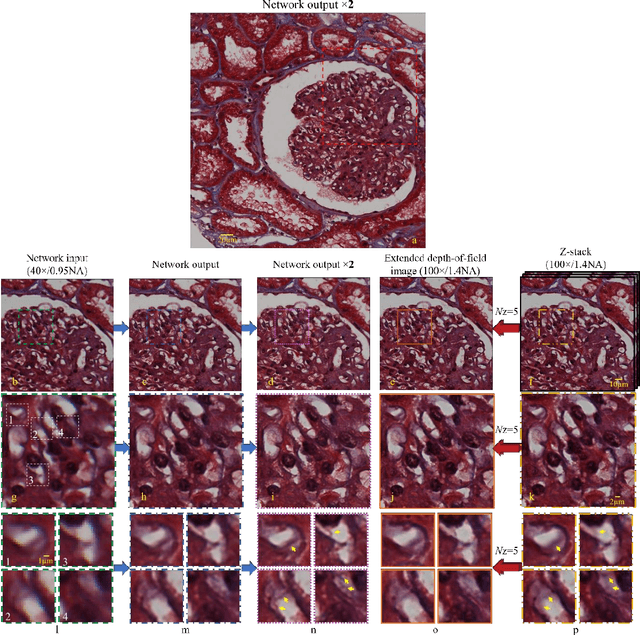
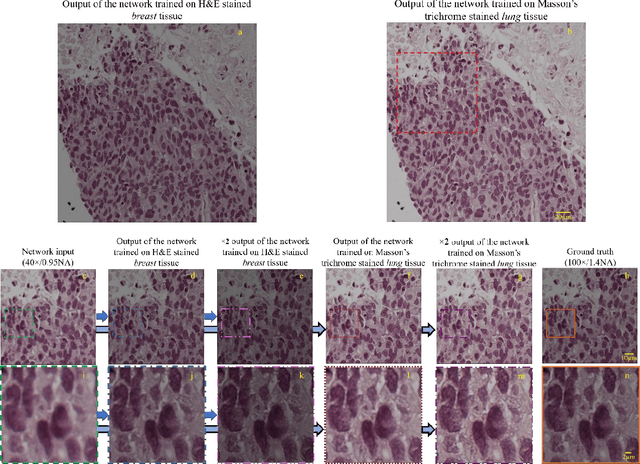
Abstract:We demonstrate that a deep neural network can significantly improve optical microscopy, enhancing its spatial resolution over a large field-of-view and depth-of-field. After its training, the only input to this network is an image acquired using a regular optical microscope, without any changes to its design. We blindly tested this deep learning approach using various tissue samples that are imaged with low-resolution and wide-field systems, where the network rapidly outputs an image with remarkably better resolution, matching the performance of higher numerical aperture lenses, also significantly surpassing their limited field-of-view and depth-of-field. These results are transformative for various fields that use microscopy tools, including e.g., life sciences, where optical microscopy is considered as one of the most widely used and deployed techniques. Beyond such applications, our presented approach is broadly applicable to other imaging modalities, also spanning different parts of the electromagnetic spectrum, and can be used to design computational imagers that get better and better as they continue to image specimen and establish new transformations among different modes of imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge