Yair Rivenson
Virtual Gram staining of label-free bacteria using darkfield microscopy and deep learning
Jul 17, 2024



Abstract:Gram staining has been one of the most frequently used staining protocols in microbiology for over a century, utilized across various fields, including diagnostics, food safety, and environmental monitoring. Its manual procedures make it vulnerable to staining errors and artifacts due to, e.g., operator inexperience and chemical variations. Here, we introduce virtual Gram staining of label-free bacteria using a trained deep neural network that digitally transforms darkfield images of unstained bacteria into their Gram-stained equivalents matching brightfield image contrast. After a one-time training effort, the virtual Gram staining model processes an axial stack of darkfield microscopy images of label-free bacteria (never seen before) to rapidly generate Gram staining, bypassing several chemical steps involved in the conventional staining process. We demonstrated the success of the virtual Gram staining workflow on label-free bacteria samples containing Escherichia coli and Listeria innocua by quantifying the staining accuracy of the virtual Gram staining model and comparing the chromatic and morphological features of the virtually stained bacteria against their chemically stained counterparts. This virtual bacteria staining framework effectively bypasses the traditional Gram staining protocol and its challenges, including stain standardization, operator errors, and sensitivity to chemical variations.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023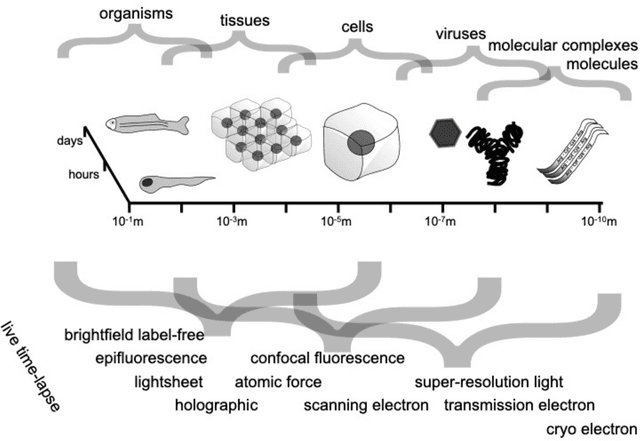
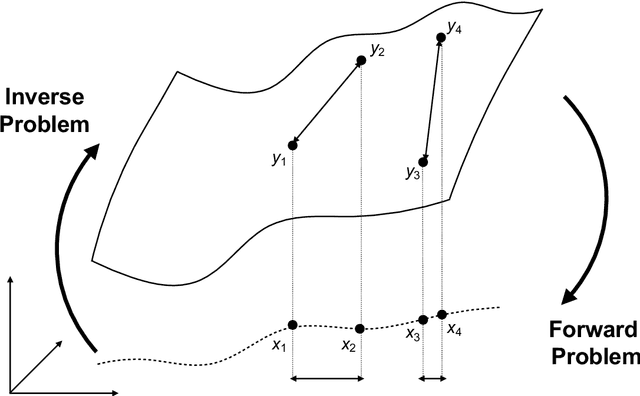
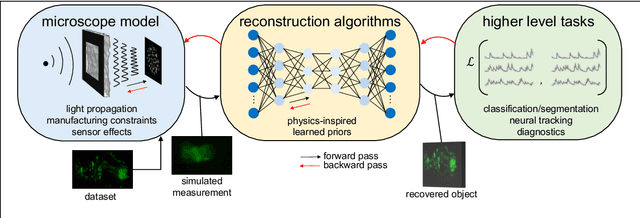
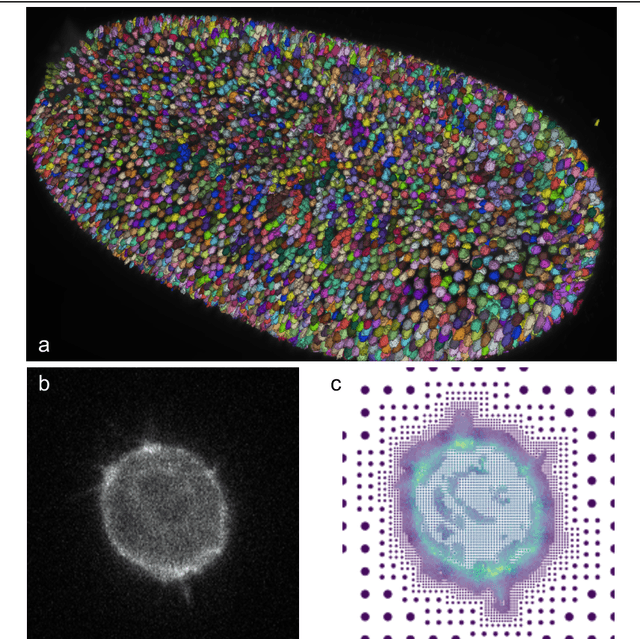
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Label-free virtual HER2 immunohistochemical staining of breast tissue using deep learning
Dec 08, 2021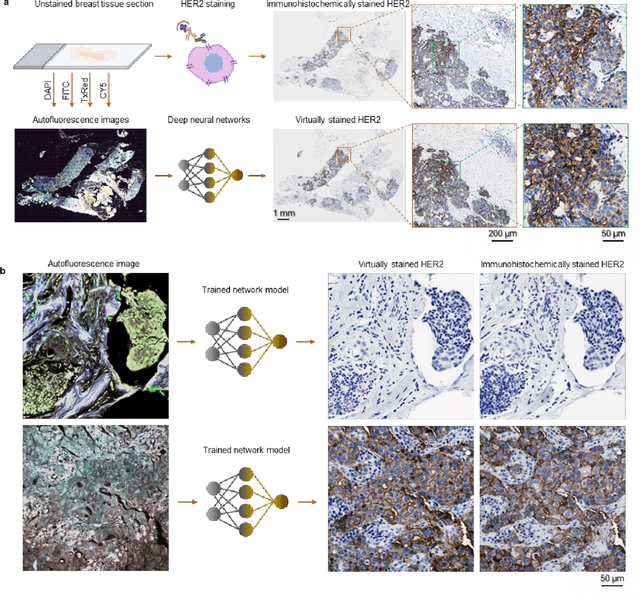
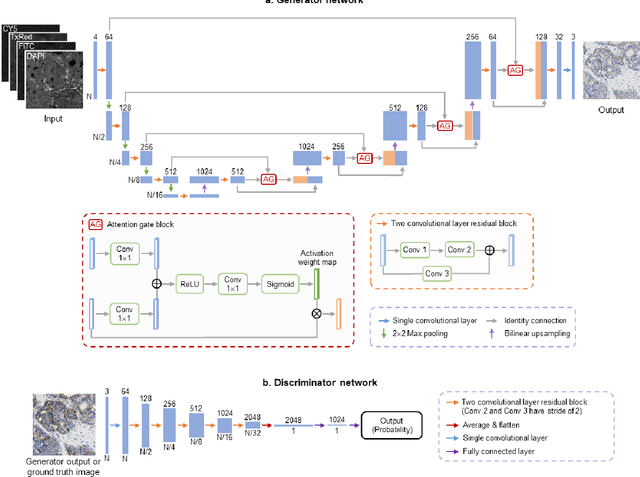
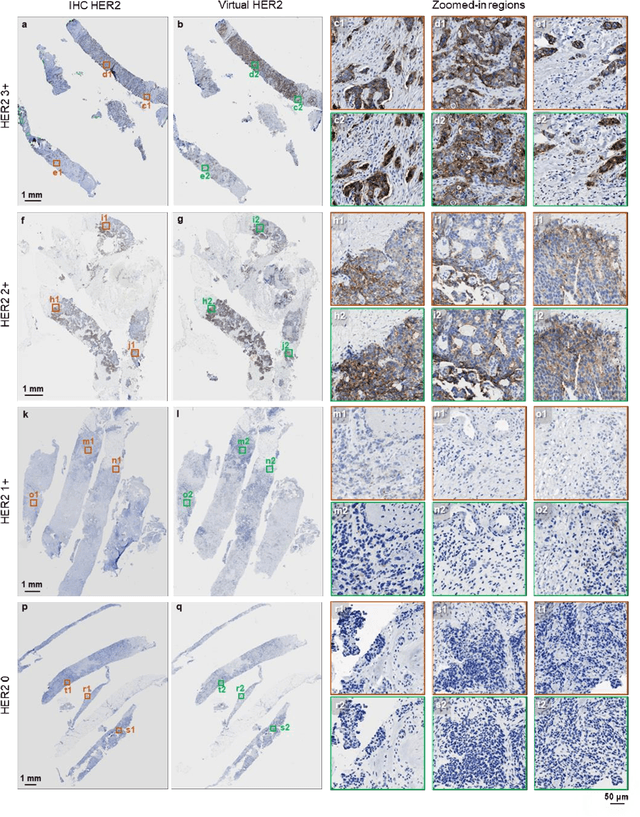
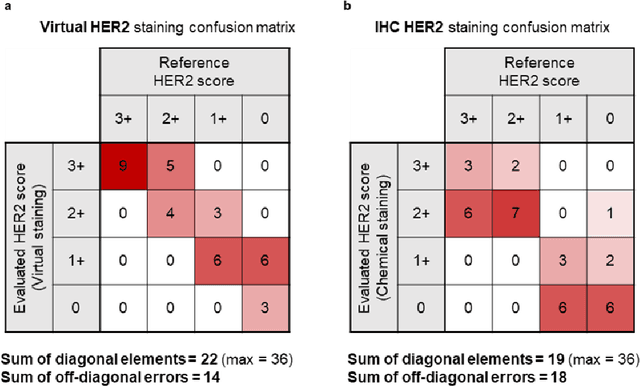
Abstract:The immunohistochemical (IHC) staining of the human epidermal growth factor receptor 2 (HER2) biomarker is widely practiced in breast tissue analysis, preclinical studies and diagnostic decisions, guiding cancer treatment and investigation of pathogenesis. HER2 staining demands laborious tissue treatment and chemical processing performed by a histotechnologist, which typically takes one day to prepare in a laboratory, increasing analysis time and associated costs. Here, we describe a deep learning-based virtual HER2 IHC staining method using a conditional generative adversarial network that is trained to rapidly transform autofluorescence microscopic images of unlabeled/label-free breast tissue sections into bright-field equivalent microscopic images, matching the standard HER2 IHC staining that is chemically performed on the same tissue sections. The efficacy of this virtual HER2 staining framework was demonstrated by quantitative analysis, in which three board-certified breast pathologists blindly graded the HER2 scores of virtually stained and immunohistochemically stained HER2 whole slide images (WSIs) to reveal that the HER2 scores determined by inspecting virtual IHC images are as accurate as their immunohistochemically stained counterparts. A second quantitative blinded study performed by the same diagnosticians further revealed that the virtually stained HER2 images exhibit a comparable staining quality in the level of nuclear detail, membrane clearness, and absence of staining artifacts with respect to their immunohistochemically stained counterparts. This virtual HER2 staining framework bypasses the costly, laborious, and time-consuming IHC staining procedures in laboratory, and can be extended to other types of biomarkers to accelerate the IHC tissue staining used in life sciences and biomedical workflow.
All-Optical Synthesis of an Arbitrary Linear Transformation Using Diffractive Surfaces
Aug 22, 2021



Abstract:We report the design of diffractive surfaces to all-optically perform arbitrary complex-valued linear transformations between an input (N_i) and output (N_o), where N_i and N_o represent the number of pixels at the input and output fields-of-view (FOVs), respectively. First, we consider a single diffractive surface and use a matrix pseudoinverse-based method to determine the complex-valued transmission coefficients of the diffractive features/neurons to all-optically perform a desired/target linear transformation. In addition to this data-free design approach, we also consider a deep learning-based design method to optimize the transmission coefficients of diffractive surfaces by using examples of input/output fields corresponding to the target transformation. We compared the all-optical transformation errors and diffraction efficiencies achieved using data-free designs as well as data-driven (deep learning-based) diffractive designs to all-optically perform (i) arbitrarily-chosen complex-valued transformations including unitary, nonunitary and noninvertible transforms, (ii) 2D discrete Fourier transformation, (iii) arbitrary 2D permutation operations, and (iv) high-pass filtered coherent imaging. Our analyses reveal that if the total number (N) of spatially-engineered diffractive features/neurons is N_i x N_o or larger, both design methods succeed in all-optical implementation of the target transformation, achieving negligible error. However, compared to data-free designs, deep learning-based diffractive designs are found to achieve significantly larger diffraction efficiencies for a given N and their all-optical transformations are more accurate for N < N_i x N_o. These conclusions are generally applicable to various optical processors that employ spatially-engineered diffractive surfaces.
Classification and reconstruction of spatially overlapping phase images using diffractive optical networks
Aug 18, 2021



Abstract:Diffractive optical networks unify wave optics and deep learning to all-optically compute a given machine learning or computational imaging task as the light propagates from the input to the output plane. Here, we report the design of diffractive optical networks for the classification and reconstruction of spatially overlapping, phase-encoded objects. When two different phase-only objects spatially overlap, the individual object functions are perturbed since their phase patterns are summed up. The retrieval of the underlying phase images from solely the overlapping phase distribution presents a challenging problem, the solution of which is generally not unique. We show that through a task-specific training process, passive diffractive networks composed of successive transmissive layers can all-optically and simultaneously classify two different randomly-selected, spatially overlapping phase images at the input. After trained with ~550 million unique combinations of phase-encoded handwritten digits from the MNIST dataset, our blind testing results reveal that the diffractive network achieves an accuracy of >85.8% for all-optical classification of two overlapping phase images of new handwritten digits. In addition to all-optical classification of overlapping phase objects, we also demonstrate the reconstruction of these phase images based on a shallow electronic neural network that uses the highly compressed output of the diffractive network as its input (with e.g., ~20-65 times less number of pixels) to rapidly reconstruct both of the phase images, despite their spatial overlap and related phase ambiguity. The presented phase image classification and reconstruction framework might find applications in e.g., computational imaging, microscopy and quantitative phase imaging fields.
Computational Imaging Without a Computer: Seeing Through Random Diffusers at the Speed of Light
Jul 14, 2021



Abstract:Imaging through diffusers presents a challenging problem with various digital image reconstruction solutions demonstrated to date using computers. We present a computer-free, all-optical image reconstruction method to see through random diffusers at the speed of light. Using deep learning, a set of diffractive surfaces are designed/trained to all-optically reconstruct images of objects that are covered by random phase diffusers. We experimentally demonstrated this concept using coherent THz illumination and all-optically reconstructed objects distorted by unknown, random diffusers, never used during training. Unlike digital methods, all-optical diffractive reconstructions do not require power except for the illumination light. This diffractive solution to see through diffusers can be extended to other wavelengths, and might fuel various applications in biomedical imaging, astronomy, atmospheric sciences, oceanography, security, robotics, among others.
Neural network-based image reconstruction in swept-source optical coherence tomography using undersampled spectral data
Mar 04, 2021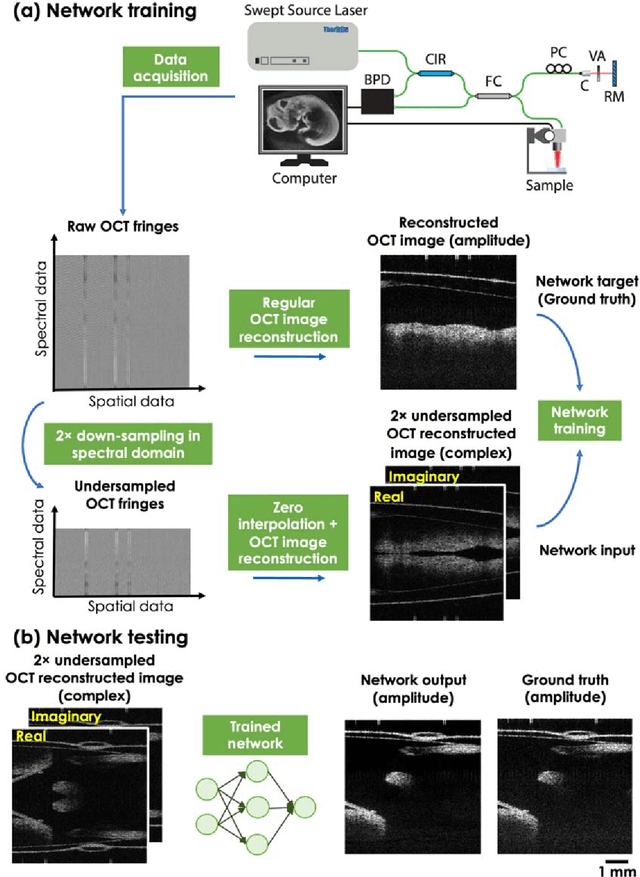
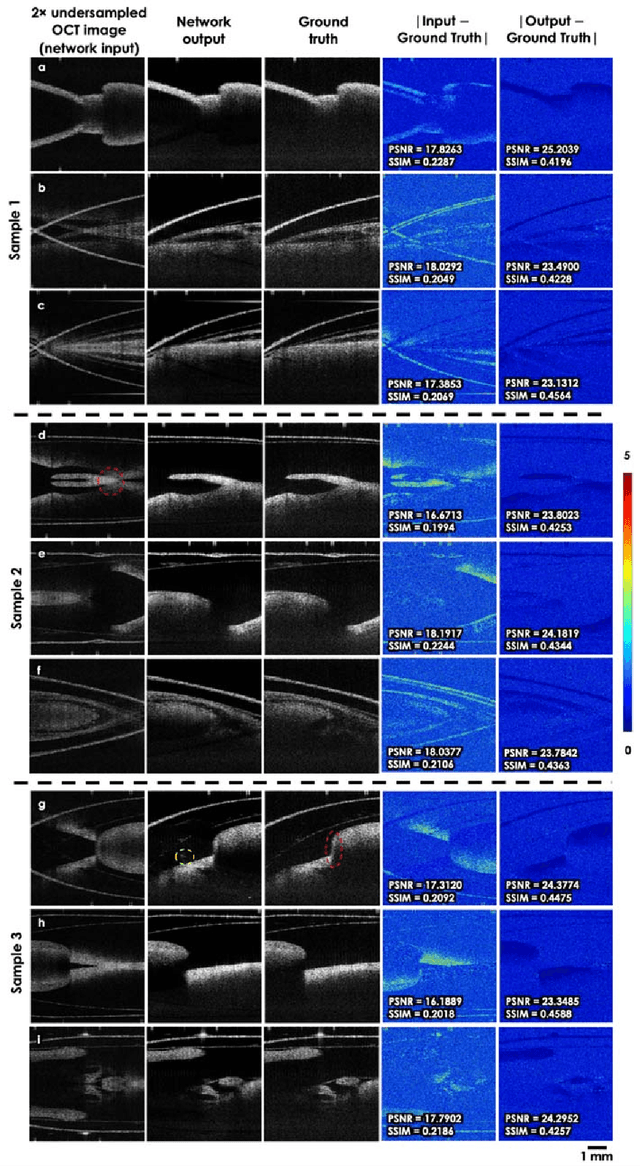
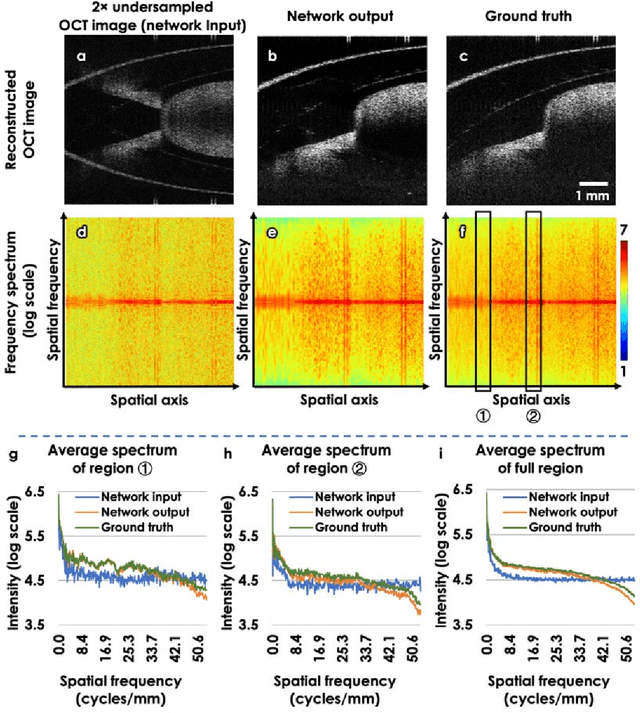
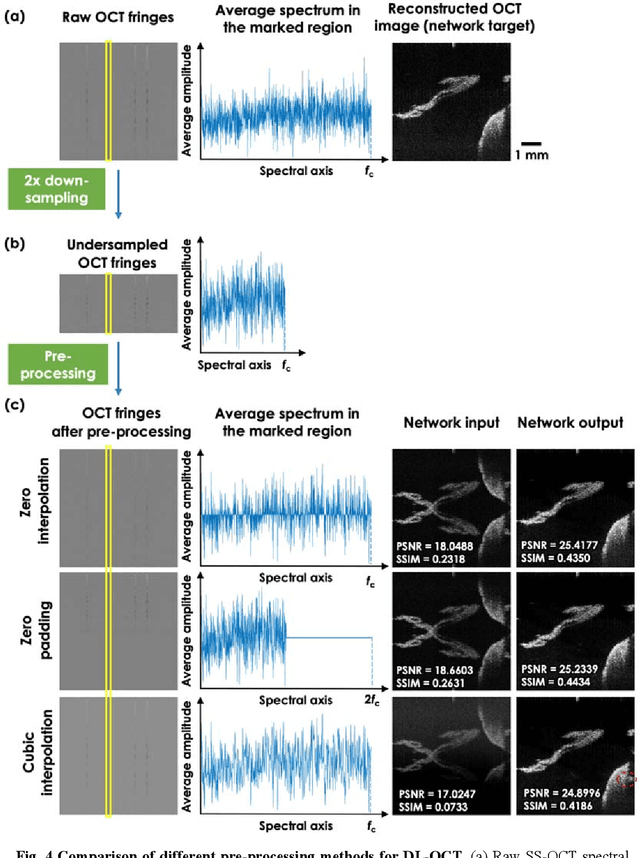
Abstract:Optical Coherence Tomography (OCT) is a widely used non-invasive biomedical imaging modality that can rapidly provide volumetric images of samples. Here, we present a deep learning-based image reconstruction framework that can generate swept-source OCT (SS-OCT) images using undersampled spectral data, without any spatial aliasing artifacts. This neural network-based image reconstruction does not require any hardware changes to the optical set-up and can be easily integrated with existing swept-source or spectral domain OCT systems to reduce the amount of raw spectral data to be acquired. To show the efficacy of this framework, we trained and blindly tested a deep neural network using mouse embryo samples imaged by an SS-OCT system. Using 2-fold undersampled spectral data (i.e., 640 spectral points per A-line), the trained neural network can blindly reconstruct 512 A-lines in ~6.73 ms using a desktop computer, removing spatial aliasing artifacts due to spectral undersampling, also presenting a very good match to the images of the same samples, reconstructed using the full spectral OCT data (i.e., 1280 spectral points per A-line). We also successfully demonstrate that this framework can be further extended to process 3x undersampled spectral data per A-line, with some performance degradation in the reconstructed image quality compared to 2x spectral undersampling. This deep learning-enabled image reconstruction approach can be broadly used in various forms of spectral domain OCT systems, helping to increase their imaging speed without sacrificing image resolution and signal-to-noise ratio.
Holographic image reconstruction with phase recovery and autofocusing using recurrent neural networks
Feb 12, 2021



Abstract:Digital holography is one of the most widely used label-free microscopy techniques in biomedical imaging. Recovery of the missing phase information of a hologram is an important step in holographic image reconstruction. Here we demonstrate a convolutional recurrent neural network (RNN) based phase recovery approach that uses multiple holograms, captured at different sample-to-sensor distances to rapidly reconstruct the phase and amplitude information of a sample, while also performing autofocusing through the same network. We demonstrated the success of this deep learning-enabled holography method by imaging microscopic features of human tissue samples and Papanicolaou (Pap) smears. These results constitute the first demonstration of the use of recurrent neural networks for holographic imaging and phase recovery, and compared with existing methods, the presented approach improves the reconstructed image quality, while also increasing the depth-of-field and inference speed.
Deep learning-based virtual refocusing of images using an engineered point-spread function
Dec 22, 2020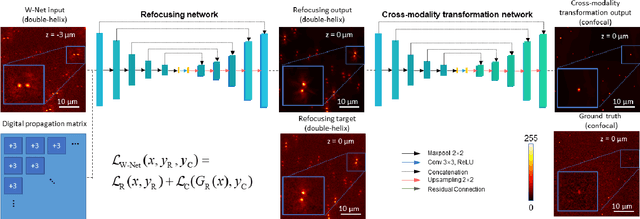
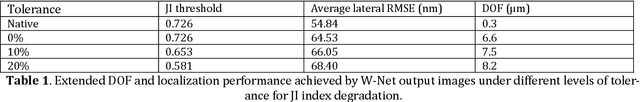
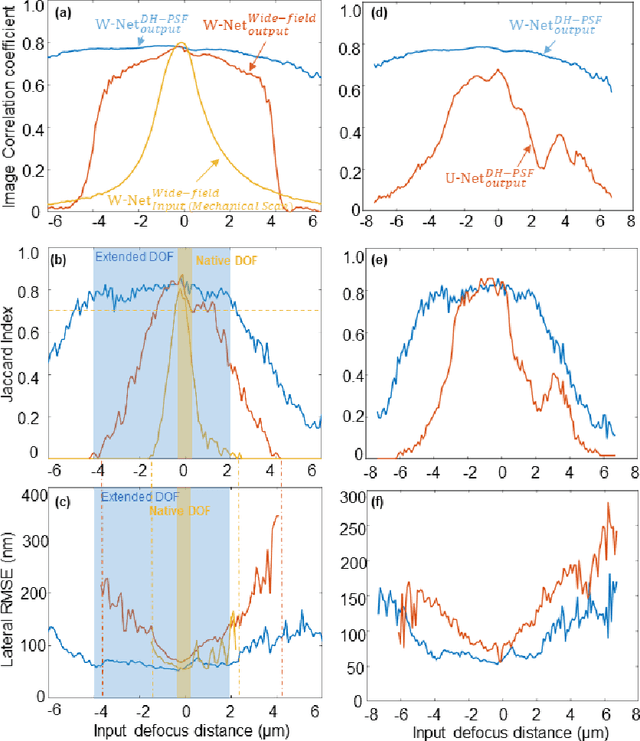
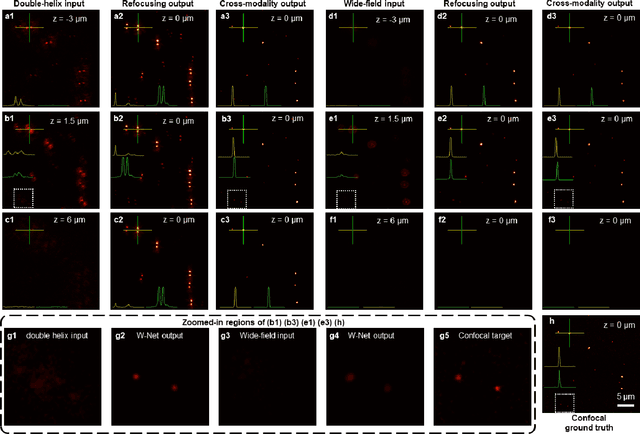
Abstract:We present a virtual image refocusing method over an extended depth of field (DOF) enabled by cascaded neural networks and a double-helix point-spread function (DH-PSF). This network model, referred to as W-Net, is composed of two cascaded generator and discriminator network pairs. The first generator network learns to virtually refocus an input image onto a user-defined plane, while the second generator learns to perform a cross-modality image transformation, improving the lateral resolution of the output image. Using this W-Net model with DH-PSF engineering, we extend the DOF of a fluorescence microscope by ~20-fold. This approach can be applied to develop deep learning-enabled image reconstruction methods for localization microscopy techniques that utilize engineered PSFs to improve their imaging performance, including spatial resolution and volumetric imaging throughput.
Neural network-based on-chip spectroscopy using a scalable plasmonic encoder
Dec 01, 2020

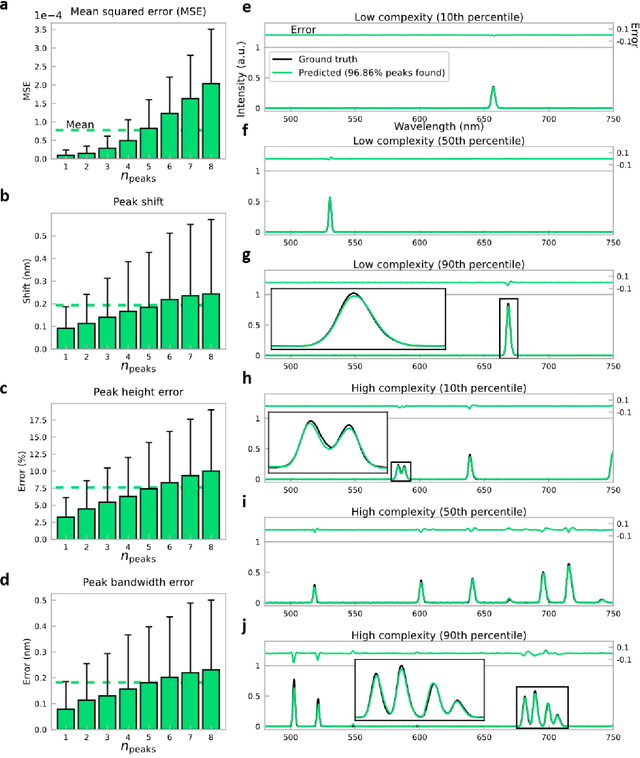
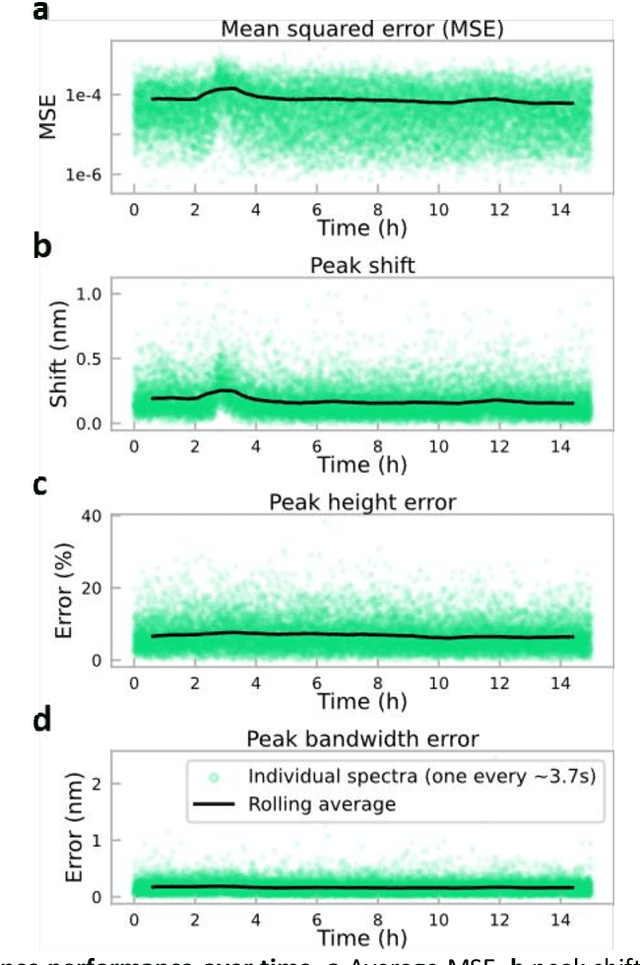
Abstract:Conventional spectrometers are limited by trade-offs set by size, cost, signal-to-noise ratio (SNR), and spectral resolution. Here, we demonstrate a deep learning-based spectral reconstruction framework, using a compact and low-cost on-chip sensing scheme that is not constrained by the design trade-offs inherent to grating-based spectroscopy. The system employs a plasmonic spectral encoder chip containing 252 different tiles of nanohole arrays fabricated using a scalable and low-cost imprint lithography method, where each tile has a unique geometry and, thus, a unique optical transmission spectrum. The illumination spectrum of interest directly impinges upon the plasmonic encoder, and a CMOS image sensor captures the transmitted light, without any lenses, gratings, or other optical components in between, making the entire hardware highly compact, light-weight and field-portable. A trained neural network then reconstructs the unknown spectrum using the transmitted intensity information from the spectral encoder in a feed-forward and non-iterative manner. Benefiting from the parallelization of neural networks, the average inference time per spectrum is ~28 microseconds, which is orders of magnitude faster compared to other computational spectroscopy approaches. When blindly tested on unseen new spectra (N = 14,648) with varying complexity, our deep-learning based system identified 96.86% of the spectral peaks with an average peak localization error, bandwidth error, and height error of 0.19 nm, 0.18 nm, and 7.60%, respectively. This system is also highly tolerant to fabrication defects that may arise during the imprint lithography process, which further makes it ideal for applications that demand cost-effective, field-portable and sensitive high-resolution spectroscopy tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge