Leveraging Computational Pathology AI for Noninvasive Optical Imaging Analysis Without Retraining
Paper and Code
Nov 19, 2024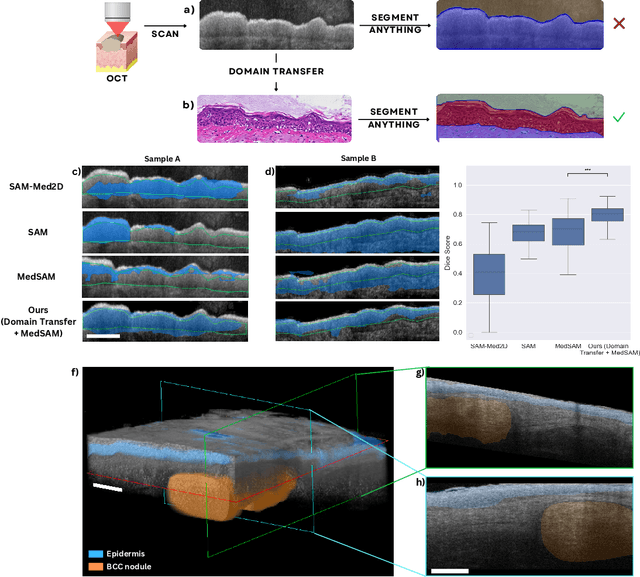
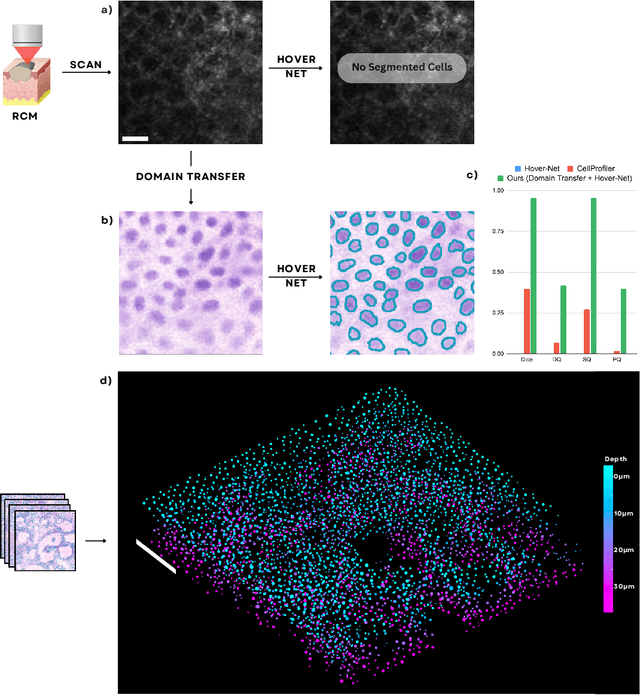
Noninvasive optical imaging modalities can probe patient's tissue in 3D and over time generate gigabytes of clinically relevant data per sample. There is a need for AI models to analyze this data and assist clinical workflow. The lack of expert labelers and the large dataset required (>100,000 images) for model training and tuning are the main hurdles in creating foundation models. In this paper we introduce FoundationShift, a method to apply any AI model from computational pathology without retraining. We show our method is more accurate than state of the art models (SAM, MedSAM, SAM-Med2D, CellProfiler, Hover-Net, PLIP, UNI and ChatGPT), with multiple imaging modalities (OCT and RCM). This is achieved without the need for model retraining or fine-tuning. Applying our method to noninvasive in vivo images could enable physicians to readily incorporate optical imaging modalities into their clinical practice, providing real time tissue analysis and improving patient care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge